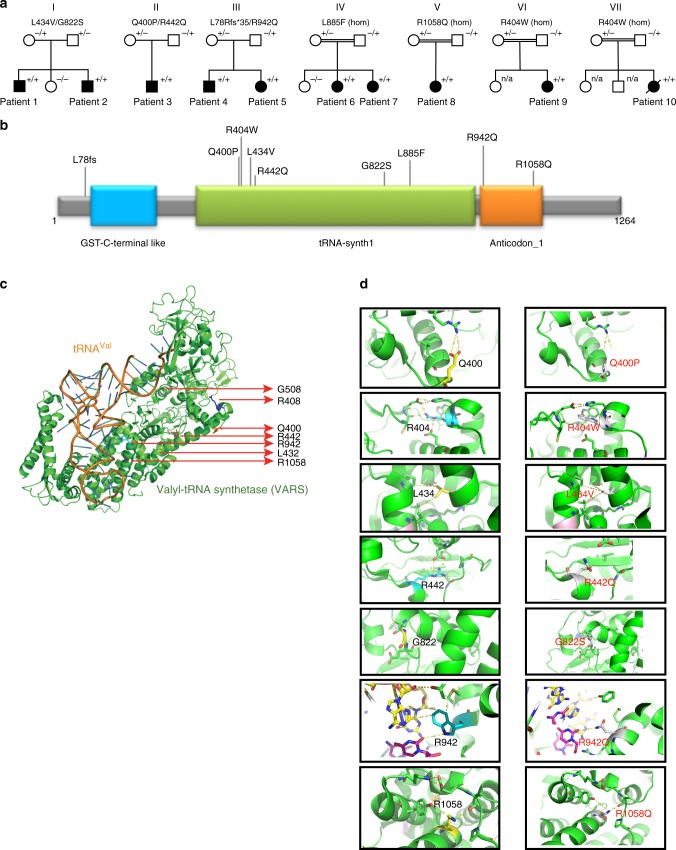
Fig. 1.
Identification of VARS variants in seven families with developmental encephalophaties and in silico predictions. a Pedigrees of the seven families diagnosed with VARS mutations. b Location of the identified VARS variants on protein level (InterPro/P26640). c Ribbon cartoon model of the Thermus thermophilus VARS-tRNA complex, highlighting the residues corresponding to those substituted in the human model. d Pair-wise comparisons between the wild-type (left) and mutant (right) residues for predicted changes in local contacts with tRNA or other amino acids. Hydrogen bonds were indicated as dotted yellow lines