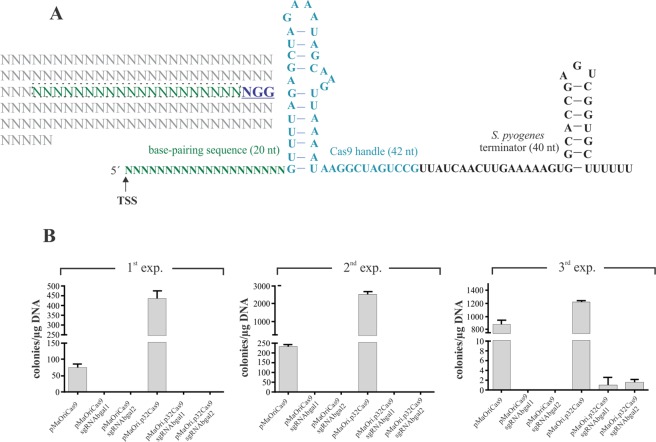
Figure 3.
sgRNA-directed Ca9 genomic cleavage is lethal to L. biflexa cells. (A) Protospacers are selected based on the presence of a 3′ NGG (PAM). Schematic representation of sgRNA sequence is also shown; the first 20–25 nucleotides, starting from the transcription start site (TSS) are variable according to the gene target, followed by a sequence folded in a hairpin-like structure responsible for Cas9 interaction. Transcription termination is dictated by the presence of a S. pyogenes intrinsic terminator. (B) pMaOriCas9, pMaOri.p32Cas9 plasmids, alone or containing different sgRNA targeting the β-galactosidase gene were used to electroporate competent L. biflexa cells, which were seeded onto EMJH plates plus spectinomycin and grown colonies were counted and normalized per µg of DNA. Different experiments are presented and for each, cell counting is presented as average plus standard deviation of 3 different plates.