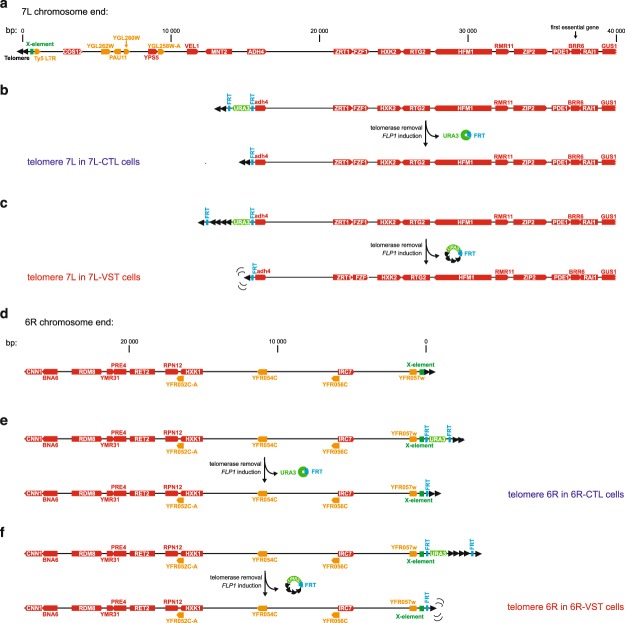
Figure 1.
Experimental system to shorten a single telomere in the cell. The chromosome end containing the 7L telomere (a) is modified in two ways. In control (CTL) cells (b), the last 15 kb of the telomere end are replaced by a construct in which a URA3 marker is flanked by two Flippase Recognition Target (FRT) sites and followed by a wild-type length telomeric tract. (c) In cells able to generate a very short telomere (VST), the URA3 marker is followed by extratelomeric repeats that inhibit the action of telomerase on telomeric repeats in cis. This results in a short terminal telomeric tract. (b,c) Upon telomerase removal and induction of flippase (encoded by FLP1), sequences between the FRT sites are excised in a circle that is diluted out upon successive cell divisions. Remaining telomeres are of wild-type length (b) or very short (c). (d) The wild type 6R chromosome end is modified to generate 6R-CTL (e) and 6R-VST (f) in a manner similar to 7L (a–c). Chromosome maps and annotations are from the Saccharomyces Genome Database and Centre for Genetic Architecture of Complex Traits website (https://www.le.ac.uk/colleges/medbiopsych/research/gact/resources/yeast-telomeres) using the sequence of strain S288C as reference. Red and orange colors correspond to genes encoding minimally characterized proteins or dubious/putative proteins, respectively. Note that subtelomeric sequences of strains W303 – used in this work - and S288C – used as reference - 7L diverge in a region spanning Ty5 to YGL262W.