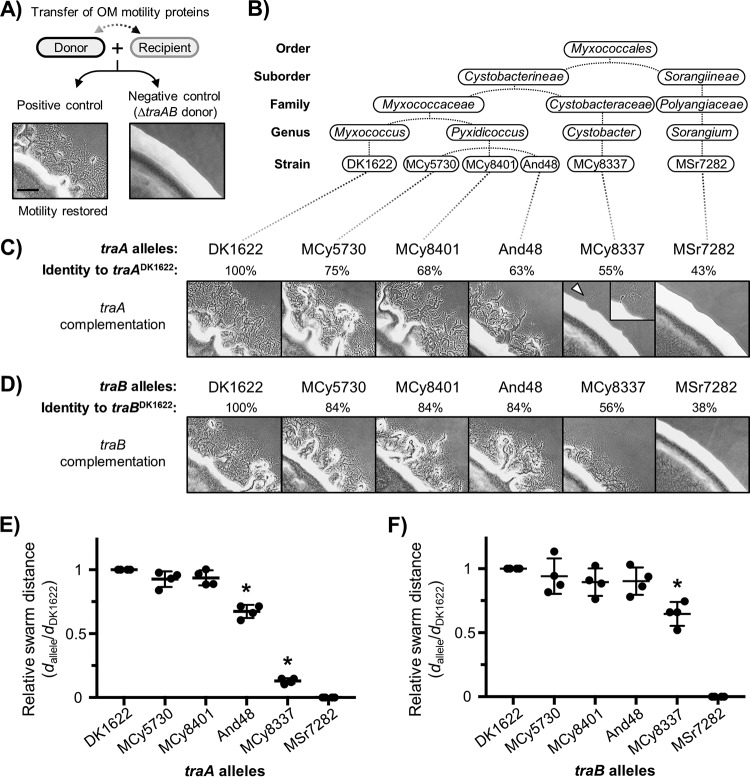
FIG 2.
Functional characterization of diverse traAB orthologs in Myxococcales. (A) Schematic of the stimulation assay that restores motility to certain mutants (recipients) by the transfer of wild-type motility proteins from nonmotile donors. A positive control (donor and recipient, with both harboring traABDK1622) and a negative control (donor lacking traAB) are shown. (B) Taxonomic origins of the traAB alleles analyzed. (C) Stimulation assays testing for functional complementation by heterologously expressing different traA alleles in isogenic ΔtraA M. xanthus strains. Each micrograph shows a mixture of donors and recipients bearing identical traA alleles. Allele names and their percentages of identical amino acids relative to traADK1622 (full length) are shown. The arrowhead highlights a small emergent flare at the edge of the colony. The inset shows an enlarged view of stimulated cells. (D) Stimulation assays testing for complementation of heterologous traB alleles in isogenic ΔtraB strains. Allele names and allele identities to traBDK1622 are indicated. Stimulation efficacy was calculated by measuring the distance (d) of the movement of emergent flares from colony edges. Data representing the relative swarming distances determined in traA and traB complementation experiments are shown in panels E and F, respectively. Four experimental replicates were done. Error bars represent standard deviations from the means. Significant differences between the DK1622 group and other groups (i.e., functionally distant alleles) are indicated by asterisks (P < 0.05 [t test]). Strain details are given in Table 2 (see also Table S1). Scale bar, 200 µm.