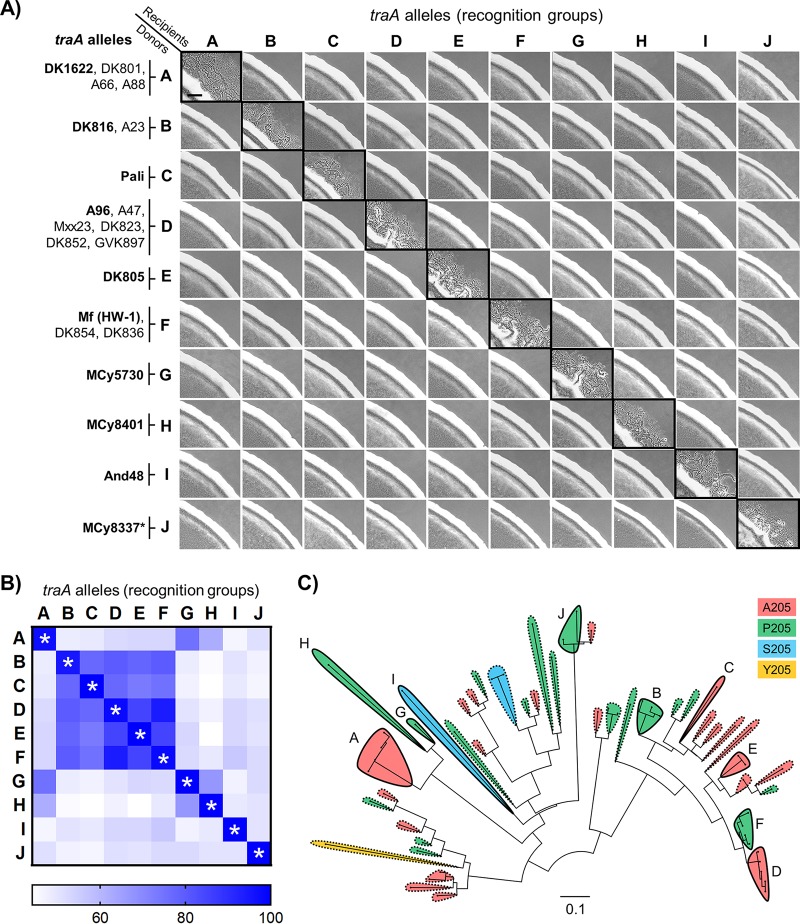
FIG 6.
Self-recognition among a wide range of myxobacteria is governed by the traA locus. (A) Stimulation assays showing specific recognition among 10 traA alleles (names are shown in bold on the left) in an isogenic set of strains. Black borders highlight 10 distinct recognition groups (groups A to J). Additional group members that have been functionally characterized (5, 10) are also listed on the left. The asterisk indicates a chimeric allele harboring VDMCy8337 (see Fig. S3 for details). Scale bar, 200 µm. (B) Pairwise plot of (%) identity among VDs of the TraA orthologs tested in panel A (asterisks indicate self-recognition). (C) Same tree as that shown in Fig. 5B, where allele names are given. Shaded areas highlight distinct recognition groups, solid lines indicate characterized recognition groups (letters indicate group names), and dashed lines show predicted recognition groups. Groups are color coded according to the specificity-determining residue at position 205. The scale bar indicates the number of substitutions per amino acid residue.