Various mechanisms have been identified by which viruses target host small RNA biogenesis pathways to achieve optimal infection outcomes. Herpes simplex virus 1 (HSV-1) is a ubiquitous human pathogen whose successful persistence in the host entails both productive (“lytic”) and latent infection. Although many HSV-1 miRNAs have been discovered and some are thought to help control the lytic/latent switch, little is known about regulation of their biogenesis. By characterizing expression of both pre-miRNAs and mature miRNAs under various conditions, this study revealed striking differences in miRNA biogenesis between lytic and latent infection and uncovered a regulatory mechanism that blocks pre-miRNA nuclear export and is dependent on viral protein ICP27 and viral DNA synthesis. This mechanism represents a new virus-host interaction that could limit the repressive effects of HSV-1 miRNAs hypothesized to promote latency and may shed light on the regulation of miRNA nuclear export, which has been relatively unexplored.
KEYWORDS: herpes simplex virus, ICP27, latency, miRNAs, nuclear export
ABSTRACT
Herpes simplex virus 1 (HSV-1) switches between two infection programs, productive (“lytic”) and latent infection. Some HSV-1 microRNAs (miRNAs) have been hypothesized to help control this switch, and yet little is known about regulation of their expression. Using Northern blot analyses, we found that, despite inherent differences in biogenesis efficiency among six HSV-1 miRNAs, all six exhibited high pre-miRNA/miRNA ratios during lytic infection of different cell lines and, when detectable, in acutely infected mouse trigeminal ganglia. In contrast, considerably lower ratios were observed in latently infected ganglia and in cells transduced with lentiviral vectors expressing the miRNAs, suggesting that HSV-1 lytic infection blocks miRNA biogenesis. This phenomenon is not specific to viral miRNAs, as a host miRNA expressed from recombinant HSV-1 also exhibited high pre-miRNA/miRNA ratios late during lytic infection. The levels of most of the mature miRNAs remained stable during infection in the presence of actinomycin D, indicating that the high ratios are due to inefficient pre-miRNA conversion to miRNA. Cellular fractionation experiments showed that late (but not early) during infection, pre-miRNAs were enriched in the nucleus and depleted in the cytoplasm, indicating that nuclear export was blocked. A mutation eliminating ICP27 expression or addition of acyclovir reduced pre-miRNA/miRNA ratios, but mutations drastically reducing Us11 expression did not. Thus, HSV-1 lytic infection inhibits miRNA biogenesis at the step of nuclear export and does so in an ICP27- and viral DNA synthesis-dependent manner. This mechanism may benefit the virus by reducing expression of repressive miRNAs during lytic infection while permitting elevated expression during latency.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is a prevalent human pathogen that establishes lifelong latency in sensory neurons following productive (“lytic”) infection in peripheral tissues (reviewed in reference 1). HSV-1 encodes at least 21 microRNAs (miRNAs) (2–8), many of which are conserved in HSV-2 (4, 9–11). Some miRNAs, together with the ∼2-kb-intron latency-associated transcripts (LAT), are the only viral gene products known to be highly expressed during latency (12). Hypothesized functions of HSV-1 miRNAs include repression of lytic gene expression during latency (3, 13–15), repression of viral replication (5, 16), antagonism of intrinsic and innate immunity (6, 17, 18), and promotion of viral replication (7). Because miRNAs function efficiently only when they are abundant (19), it is important to understand regulation of miRNA expression.
miRNAs are derived from transcripts (pri-miRNAs) with embedded stem-loops. Typically, Drosha, a nuclear RNase, cleaves pri-miRNAs into smaller hairpin-shaped RNAs known as pre-miRNAs. Exportin 5 then binds pre-miRNAs in a Ran-GTP-dependent manner and exports them into the cytoplasm, where they are further cleaved by another RNase, Dicer, releasing small RNA duplexes. One strand of each duplex is subsequently loaded into an RNA-induced silencing complex containing the Argonaute (Ago) protein, which targets the complex to complementary mRNAs (reviewed in references 19 and 20).
HSV-1 lytic infection and latent infection display different viral miRNA expression profiles. During lytic infection, miR-H1 and miR-H6 appear to be the most abundantly expressed (4, 12, 13). However, during latency, miR-H1 is poorly expressed whereas miR-H2, miR-H4-5p, miR-H4-3p, and miR-H5 to miR-H8 are abundant (8, 12, 21–23). Except for miR-H6, these miRNAs are located within the LAT transcription unit, suggesting that they are cleavage products of the primary 8.3-kb LAT.
Most of our knowledge about HSV-1 miRNA expression has come from deep sequencing and stem-loop reverse transcription-quantitative PCR (qRT-PCR) analyses, which provide no information regarding miRNA precursors. In contrast, Northern blot hybridization (Northern) permits detection of both pre-miRNA and mature miRNAs with high specificity. Starting with the initial reports of HSV miRNAs (2, 3, 9, 10, 12, 15, 24, 25), Northern results indicated an interesting phenomenon of high expression of viral pre-miRNAs but low expression of their mature miRNAs during lytic infection. Specifically, for HSV-1, pre-miR-H1 to pre-miR-H6 are detected readily, but their mature counterparts are detected only weakly if at all (2, 3, 12, 15), in contrast to what is seen with most cellular miRNAs, such as let-7a (12). This is also true for HSV-2 miRNAs analyzed by Northern (9, 10, 24, 25). This mechanism of limited miRNA expression despite high pre-miRNA expression might operate to restrict the functions of these miRNAs during lytic infection.
To understand this phenomenon, we conducted a series of Northern-based experiments. The results not only revealed distinct HSV-1 miRNA expression patterns in cell culture and mouse ganglia but also uncovered a regulatory mechanism that inhibits pre-miRNA nuclear export.
RESULTS
High ratios of HSV-1 pre-miRNAs to miRNAs during lytic infection.
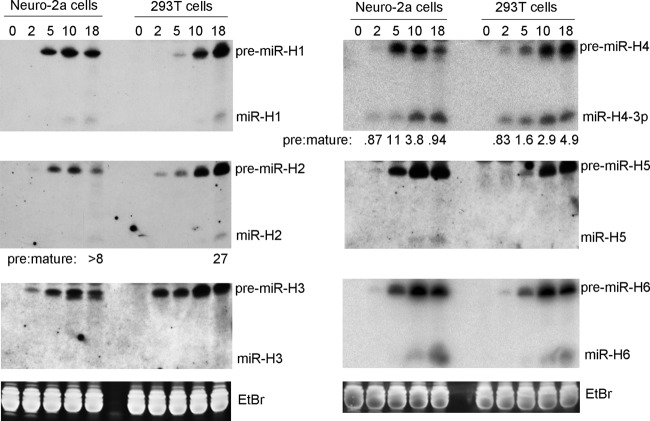
Because Northern analyses had been performed previously for HSV-1 miRNAs only in African green monkey kidney (Vero) cells, we analyzed expression of miR-H1 to miR-H6 using two additional cell lines, human embryonic kidney (293T) cells and mouse neuroblastoma (Neuro-2a) cells, following infection at multiplicities of infection (MOI) of 10 and 30, respectively, by HSV-1 wild-type (WT) strain KOS. These miRNAs (and the corresponding pre-miRNAs) are expressed as late gene products in Vero cells from unknown promoters (deletion of the LAT promoter does not affect their expression during lytic infection as it does during latency) (12). In addition to species detected by previous Northern analyses in Vero cells, including pre-miR-H1 to pre-miR-H6 and mature miR-H1, miR-H2, and miR-H6, we additionally detected mature miR-H4-3p in both 293T and Neuro-2a cells and miR-H5 in Neuro-2a cells (Fig. 1). All 6 pre-miRNAs became detectable by 5 h postinfection (hpi) and abundant at 10 and 18 hpi in both cell lines. However, mature miR-H3 was not detected in either cell line, consistent with its very low level of expression (<1 molecule/cell) as measured by qRT-PCR of <40-nucleotide (nt) RNA in Vero cells (12). Mature miR-H1, miR-H2, miR-H5, and miR-H6 were detectable at late times and at much lower abundances than their precursors. miR-H4 was detectable as early as 2 hpi, as previously observed by qRT-PCR in Vero cells (12), and eventually reached a level that was similar to that seen with pre-miR-H4 in Neuro-2a cells but still substantially less than that of pre-miR-H4 in 293T cells. Taking the results together, despite slight differences observed among different miRNAs and cell lines, there were generally high pre-miRNA/mature miRNA (pre/mature) ratios in both 293T and Neuro-2a cells, as shown here, and in Vero cells, as shown previously (2, 4, 12). Thus, this phenomenon was observed in multiple species and cell types.
FIG 1.
HSV-1 pre-miRNA and mature miRNA expression in Neuro-2a and 293T cells. Neuro-2a and 293T cells were infected with HSV-1 strain KOS at MOIs of 30 and 10, respectively. At various times postinfection, cells were harvested and small RNAs (<200 bases) were purified. Following polyacrylamide gel electrophoresis, RNAs were transferred to membranes and probed for miRNA expression by Northern blot hybridization. Cell line names and times of cell harvest (in h) are indicated at the top of the Northern images. Positions of miRNA species are labeled to the right of the images. At the bottom are ethidium bromide (EtBr) staining signals from the gels at ∼80 bases showing uniform loading for the images above them. Pre/mature ratios calculated following band quantification are shown below the images for miR-H2 and miR-H4. This experiment was performed twice. Similar results were also obtained for the specific time points indicated in Fig. 2 (see also Fig. S1).
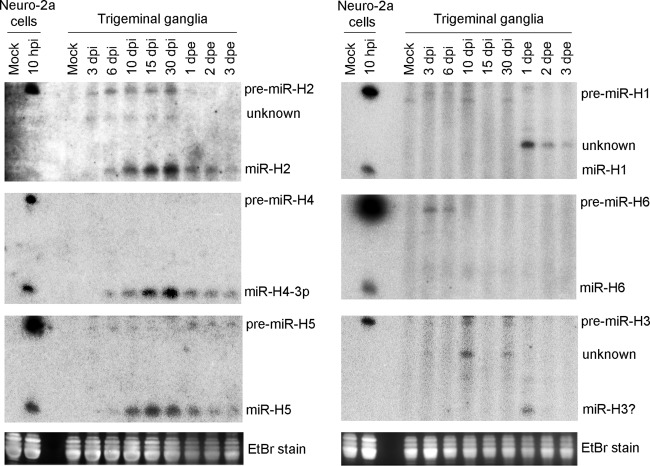
HSV-1 pre-miRNA and mature miRNA expression in trigeminal ganglia at different times after infection or after reactivation from latency. Mice were mock infected or infected on the cornea with 2 × 105 PFU of HSV-1 strain KOS per eye. At various days postinfection (dpi), trigeminal ganglia were collected. At 30 dpi, some ganglia were explanted and cultivated for various days postexplantation (dpe) before the ganglia were harvested for RNA. Sixteen ganglia were pooled for each condition, and small RNAs (<200 bases) were isolated and analyzed by Northern blot hybridization. RNAs purified from 293T cells that had been mock infected or infected for 18 h were run alongside (see last two lanes) the RNAs from ganglia to provide markers. The sources of RNAs and time points are indicated at the tops of each series (left or right) of Northern images. Positions of miRNA species are indicated to the right of the images. Ethidium bromide signals at ∼80 bases from the gels used for each series of Northern images are displayed at the bottom. An independent experiment is shown in Fig. 1. Download FIG S1, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2019 Pan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential miRNA biogenesis in acutely and latently infected mouse trigeminal ganglia.
To investigate whether high pre/mature ratios also occur in vivo, we performed two independent experiments (Fig. 2; see also Fig. S1 in the supplemental material) in which we infected mice on the cornea with 2 × 105 PFU/eye of HSV-1 and analyzed miRNA expression in trigeminal ganglia on various days postinfection (dpi) by Northern blotting. We also explanted some ganglia at 30 dpi and cultivated them for 1 to 3 days to reactivate the virus before collecting the tissues for RNA analysis. miR-H2, miR-H4, and miR-H5 showed one expression pattern: Their mature miRNAs were readily detected after 6 dpi, with levels steadily increasing through 30 dpi and then decreasing during reactivation (Fig. 2; see also Fig. S1). This is consistent with previous qRT-PCR data (12, 22, 26). However, during establishment and maintenance of latency (6 to 30 dpi), pre-miR-H4 was undetectable and pre-miR-H2 and H5 were much less abundant than their mature forms (Fig. 2; see also Fig. S1). The miRNAs that are most abundant during lytic infection, miR-H1 and miR-H6, showed a different pattern. At the height of acute ganglionic infection (3 dpi), pre-miR-H1, but not miR-H1, was detected weakly, and neither was detected subsequently (Fig. 2; see also Fig. S1), consistent with previous qRT-PCR results (4, 12, 22). Pre-miR-H6, but not mature miR-H6, was clearly detectable at 3 and 6 dpi, but neither pre-miR-H6 nor mature miR-H6 was detected later (Fig. 2; see also Fig. S1). Our inability to detect miR-H6 was not consistent with previous qRT-PCR results that showed high miR-H6 expression levels in both acutely infected ganglia and latently infected ganglia (3, 12, 22). Insufficient sensitivity of our Northern analyses might explain this discrepancy. Finally, we did not reproducibly detect bands corresponding to pre-miR-H3 or mature miR-H3 that were clearly distinct from background in mouse ganglia, which was consistent with undetectable levels in previous qRT-PCR results obtained from <40-nt RNA (12) but not with reported high levels in unfractionated RNA (22). We suspect that the latter result was due to the presence of longer species contributing to miR-H3 qRT-PCR signals. Taking the results together, during establishment and maintenance of latency, mature miRNAs, whenever detectable, were always more abundant than pre-miRNAs (i.e., low pre/mature ratios), whereas at the height of acute infection (3 dpi), no mature miRNA was consistently detected despite multiple pre-miRNAs being detected (high pre/mature ratios), highlighting the contrast between acute infection and latent infection in miRNA biogenesis.
FIG 2.
HSV-1 pre-miRNA and mature miRNA expression in trigeminal ganglia at different times after infection or after reactivation from latency. Mice were mock infected or infected on the cornea with 2 × 105 PFU of HSV-1 strain KOS per eye. At the various days postinfection (dpi) indicated, trigeminal ganglia were collected for immediate RNA isolation. Additionally, at 30 dpi, some ganglia were explanted and cultivated for various days postexplantation (dpe) before RNA was isolated. Sixteen ganglia were pooled for each condition, and small RNAs (<200 bases) were isolated and analyzed by Northern blot hybridization. RNAs purified from Neuro-2a cells that had been mock infected or infected with HSV-1 strain KOS for 10 h were run alongside (first two lanes) the RNAs from ganglia to provide markers. The sources of RNAs and time points are indicated at the tops of each series (left and right, respectively) of Northern images. Positions of miRNA species are indicated to the right of the images. Ethidium bromide signals at ∼80 bases from the gels used for each series of Northern images are displayed at the bottom. Results of an independent experiment are shown in Fig. S1.
Lower pre/mature ratios in lentivirus-transduced cells than during HSV-1 lytic infection.
We next asked whether the high pre/mature ratios seen during lytic infection might have been due simply to a lack of sufficient time to complete biogenesis. To determine how quickly mature miRNAs could be expressed in cells, we employed an inducible lentiviral transduction system. We cloned sequences encoding miR-H2 or miR-H3/H4 pre-miRNAs along with flanking sequences into the pTRIPZ vector, in which inducible gene expression is driven by a tetracycline-responsive promoter (sequences encoding pre-miR-H3 and -H4 were cloned as one fragment as they are separated by only 129 bp). We then produced lentiviruses and used them to stably transduce 293T and Neuro-2a cells. We monitored miRNA expression following doxycycline (Dox) addition to the transduced cells. The three miRNAs differed greatly in their final pre/mature ratios, with miR-H4 having the lowest and miR-H3 the highest in both the 293T-derived and Neuro-2a-derived cell lines (Fig. 3), suggesting important inherent differences in biogenesis efficiency among them. Nevertheless, the transduced cells always exhibited more rapid and efficient miRNA biogenesis than that seen in HSV-1-infected cells (Fig. 1). For example, the miR-H2 and miR-H4 pre/mature ratios seen at 5 h after addition of Dox were already lower than those seen at 18 h after HSV-1 infection, and mature miR-H3 was detectable in transduced cells but undetectable in infected cells despite the high pre-miR-H3 levels in both settings. These results argue for a regulatory mechanism impeding accumulation of mature miRNAs during lytic infection.
FIG 3.
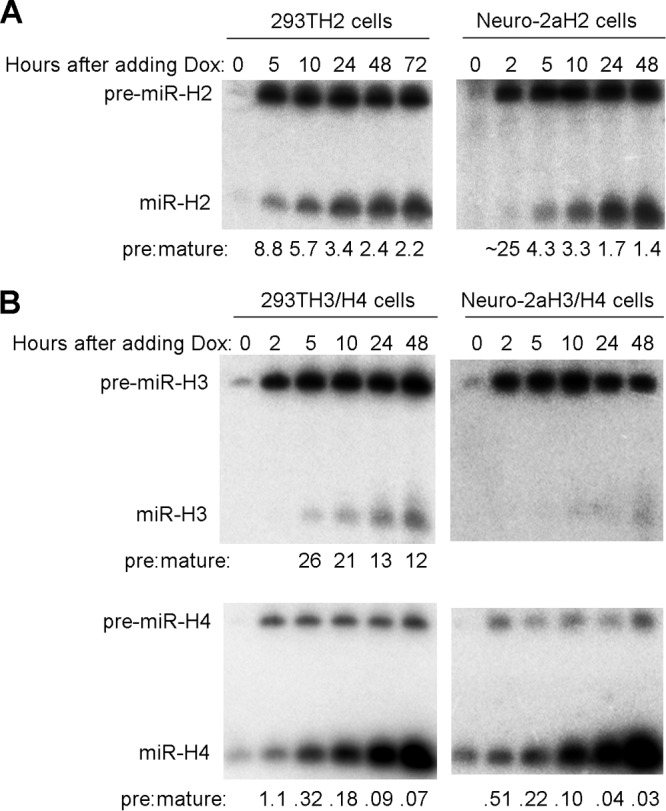
Conversion of pre-miRNA to mature miRNA during induced expression following lentiviral transduction. (A) 293T and Neuro-2a cells were stably transduced with an inducible pre-miR-H2 expressing lentivirus, resulting in 293TH2 and Neuro-2aH2 cell lines. After 1 μg/ml of Dox was added, cells were harvested at different times and analyzed by Northern blot hybridization for miR-H2 expression. Cell line names and time points are indicated at the tops of the images. Positions of miRNA species are labeled to the left. Ratios of band intensities of pre-miRNAs to those of miRNAs are shown at the bottom. The experiment using 293TH2 cells was performed twice; the experiment using Neuro-2aH2 cells was performed once. (B) As described for panel A, but the 293TH3/H4 and Neuro-2aH3/H4 cell lines were created by transduction using a lentivirus expressing miR-H3 and miR-H4, and miR-H3 and H4 expression levels were analyzed. These experiments were repeated with cells constructed using different lentivirus constructs expressing miR-H3 and/or miR-H4.
High pre/mature ratios are due to inefficient pre-miRNA conversion to mature miRNA rather than to mature miRNA destabilization.
Two hypotheses could explain the high pre/mature miRNA ratios: (i) block of pre-to-mature conversion and (ii) expedited degradation of mature miRNA. To investigate these hypotheses, we analyzed miRNA stability following addition of a transcription inhibitor, actinomycin D (ActD). In transduced 293T cells, at 3 days following Dox addition, mature miR-H2, miR-H3, and miR-H4 were all stable for 8 h following ActD addition, but the corresponding pre-miRNAs were all much less stable (Fig. 4A and B). In HSV-1-infected cells, when ActD was added at 2 hpi, pre-miR-H2 and pre-miR-H4 levels decreased substantially over the next 8 h (Fig. 4C; see also Fig. S2). However, when ActD was added at 8 hpi, pre-miR-H2 levels did not decrease over the 8 h that followed, and pre-miR-H4 and pre-miR-H1 levels decreased by only ∼2-fold during this time frame (Fig. 4D; see also Fig. S2). Following ActD addition at either 2 or 8 hpi, the levels of the corresponding mature miRNAs remained stable or even increased, with the possible exception of the level of miR-H4, which appeared to decrease and then become stable following ActD addition at 8 hpi (Fig. S2). These results suggest that inefficient conversion of pre-miRNA to mature miRNA, rather than mature miRNA instability, accounts for high pre/mature ratios and that pre-miRNAs become more stable over the course of infection, consistent with interference with pre-miRNA conversion to miRNA late in infection.
FIG 4.
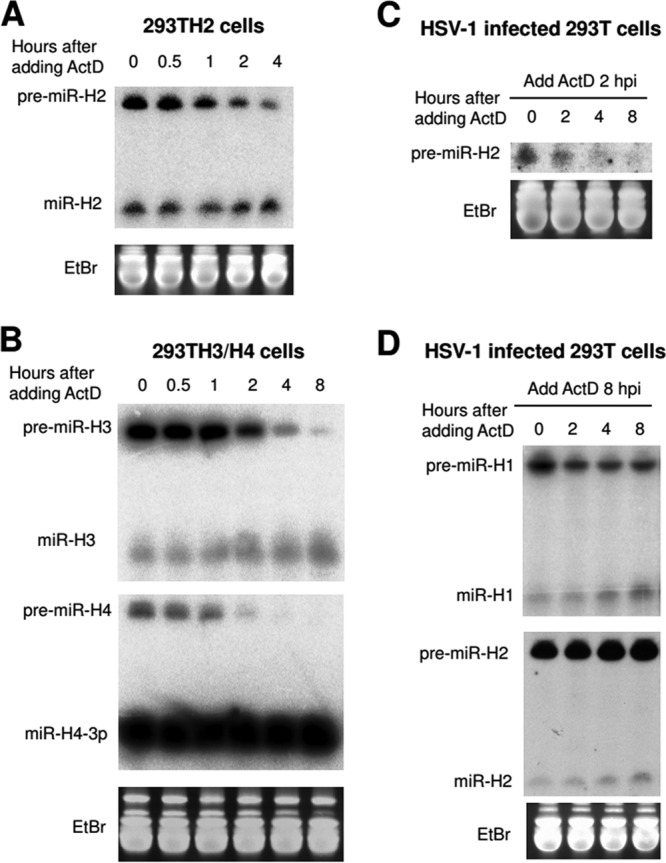
Stability of pre-miRNAs and miRNAs in lentivirus-transduced cells and HSV-1-infected cells. (A) miR-H2 expression from 293TH2 cells was induced by adding 1 μg/ml of Dox for 3 days. Then, 1 μg/ml of ActD was added to inhibit transcription. Cells were harvested at the times indicated at the top of the image and were analyzed by Northern blot hybridization for miR-H2 expression. Uniform loading on the gel used for the Northern blot hybridization was demonstrated by ethidium bromide staining (shown at the bottom). This experiment was performed once. (B) As described for panel A, but 293TH3/H4 cells were used and the levels of expression of miR-H3 (upper panel) and miR-H4 (lower panel) were analyzed. This experiment was repeated with cells constructed using different lentivirus vectors expressing miR-H3 and/or miR-H4. (C) 293T cells were infected with HSV-1 strain KOS (MOI = 10). At 2 hpi, 1 μg/ml ActD was added, and cells were harvested at the times indicated at the top of the images and analyzed by Northern blot hybridization for miR-H2 expression. The upper image shows pre-miR-H2 expression (mature miR-H2 was not detectable at that time point). The bottom images show the ethidium bromide-stained gel at ∼80 bases. (D) As described for panel C, but ActD was added at 8 hpi, and miR-H1 (upper panel) and miR-H2 (lower panel) were analyzed. Positions of miRNA species are indicated to the left. The experiments shown in panels C and D were performed twice; additionally, results for miR-H4 are shown in Fig. S2.
Stability of pre-miRNAs and miRNAs in HSV-1-infected cells. 293T cells were infected with HSV-1 strain KOS (MOI = 10). At 2 (left panel) or 8 (right panel) hpi, 1 μg/ml ActD was added, and cells were harvested at the times indicated at the top of the images. miR-H4 was analyzed by Northern blot hybridization. Results of experiments examining miR-H2 are shown in Fig. 4C and D. Download FIG S2, PDF file, 0.4 MB (421.3KB, pdf) .
Copyright © 2019 Pan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
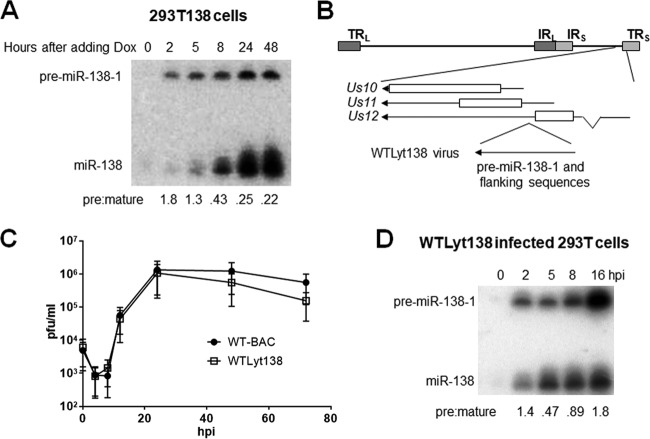
A host miRNA expressed from a recombinant HSV-1 exhibited inefficient pre-miRNA processing during lytic infection.
We wondered whether this impairment of biogenesis is specific to viral miRNAs. The effects of HSV infection on endogenous or transduced miRNAs are difficult to study due to the already high levels of most mature host miRNAs and the shutoff of most host transcription (reviewed in reference 27). To avoid these issues, we expressed miR-138 as an HSV-1 gene product. This miRNA was chosen because its low level of endogenous expression in 293T cells (28) allows easy detection of its exogenous expression from a virus. In an initial experiment, we cloned pre-miR-138-1 and flanking sequences into the inducible lentiviral transduction system described above to assess the intrinsic biogenesis efficiency of this miRNA. Upon Dox addition to miR-138-expressing 293T cells, pre-miR-138-1 levels were nearly constant from 2 to 48 h, but mature miR-138 levels increased steadily, resulting in pre/mature ratios of about 0.25 by 24 h (Fig. 5A), indicating that the biogenesis of miR-138 is intrinsically efficient. We then modified the HSV-1 genome to insert the same pre-miR138-1 and flanking sequences between the Us11 and Us12 open reading frames such that miR-138 would be encoded within the Us12 3′ untranslated region (UTR) and Us11 5′ UTR and thus would be expressed from both the Us12 immediate early and Us11 late promoters (Fig. 5B). The resulting recombinant HSV-1, designated WTLyt138, showed replication kinetics in Vero cells indistinguishable from those of the corresponding bacterial artificial chromosome (BAC)-derived WT parental virus (WT-BAC) (Fig. 5C) and showed no more than a modest defect in Us11 expression (data not shown) which should not affect miRNA biogenesis, because deletion of Us11 has little if any impact on miRNA biogenesis (see below). As expected for transcription from both immediate early and late promoters, pre-miR-138-1 expression from WTLyt138 virus appeared to be biphasic, with initial expression at 2 hpi followed by a decrease in the level of expression at 5 hpi and then an increase again after 5 hpi (Fig. 5D). Pre-miRNA conversion to mature miRNA was efficient during the first phase, with a pre/mature ratio of 0.47 at 5 hpi. After 5 hpi, however, mature miR-138 levels increased only slightly despite dramatic increases in pre-miR-138-1 levels, resulting in a 4-fold-higher pre/mature ratio at 16 hpi than at 5 hpi, arguing that biogenesis of a host miRNA was also impeded late during lytic infection.
FIG 5.
miR-138 expression in lentivirus-transduced cells and in cells infected with recombinant HSV-1 expressing miR-138. (A) 293T cells were stably transduced with an inducible pre-miR-138-1-expressing lentivirus, resulting in the 293T138 cell line. After 1 μg/ml of Dox was added, cells were harvested at the times indicated at the top of the image and analyzed by Northern blot hybridization for miR-138 expression. Pre/mature ratios calculated following band quantification are shown at the bottom. This experiment was performed twice. (B) Genomic location of inserted pre-miR-138-1-expressing sequences. The HSV-1 genome is depicted as a horizontal line at the top with long repeat sequences (TRL and IRL) and short repeat sequences (IRS and TRS), which are shown as dark and light gray boxes, respectively. An expanded view of the insertion location is shown below the horizontal line, with bars representing protein coding sequences, arrows representing transcripts and gene names provided to the left. As shown at the bottom, pre-miR-138-1 and flanking sequences were inserted between coding sequences of Us11 and Us12, resulting in a recombinant virus designated WTLyt138. (C) Replication kinetics in Vero cells (MOI = 0.02). Each point represents the average of titers from three biological replicates (the experiment was performed three times), and the vertical bars represent standard deviations. (D) 293T cells were infected with WTLyt138 (MOI = 5). The image shows an autoradiogram of the Northern blot of RNAs isolated from the cells at the hours postinfection (hpi) indicated at the top of the panel, hybridized for miR-138. Positions of pre-miR-138-1 and miR-138 are indicated to the left. Pre/mature ratios calculated following band quantification are shown at the bottom. This experiment was performed three times; additionally, similar results were obtained at two time points in Fig. 7F.
Pre-miRNA nuclear export is blocked during HSV-1 lytic infection.
To understand the mechanism of this inefficient biogenesis, we first asked whether infection changed levels of key factors involved in miRNA biogenesis and found that Drosha, Exportin 5, and Dicer were not downregulated during HSV-1 infection (Fig. S3), indicating that changes in the expression levels of these proteins did not explain inefficient miRNA biogenesis.
HSV-1 infection does not reduce expression of various proteins involved in miRNA biogenesis. 293T and Vero cells were infected at an MOI of 10 and Neuro-2a cells were infected at an MOI of 30 with HSV-1 strain KOS for the times indicated at the top of the panel. The proteins indicated to the left were analyzed by Western blotting. The experiment using Vero and 293T cells was performed multiple times. The experiment using Neuro-2a cells was performed once. Download FIG S3, PDF file, 0.2 MB (255.1KB, pdf) .
Copyright © 2019 Pan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We then analyzed miRNA expression in nuclear and cytoplasmic fractions of WTLyt138-infected cells. The biphasic expression of miR-138 from this virus facilitated analysis of this experiment. Successful fractionation was achieved and resulted in clean separation of U1 small nuclear RNA from lysine-tRNA (Fig. 6). At 5 hpi, pre-miR-138-1 was found to be present at equivalent levels in the two fractions. However, at 12 hpi, there was much more pre-miR-138-1 in the nuclear fraction than in the cytoplasmic fraction, indicating inefficient nuclear export after 5 hpi. Also, much larger fractions of pre-miR-H1 and -H6 were found in the nucleus than in the cytoplasm at 12 hpi (Fig. 6). Similar results were observed with pre-miR-H6 and mature miR-H6 in WT-infected cells (Fig. S4). We found no evidence of impaired Dicer cleavage as judged by the relatively low pre/mature ratios in the cytoplasmic fractions at 5 hpi for miR-138 and at 12 hpi for both miR-138 and miR-H6 (Fig. 6; see also Fig. S4). Therefore, we conclude that HSV-1 infection impedes miRNA biogenesis by blocking nuclear export.
FIG 6.
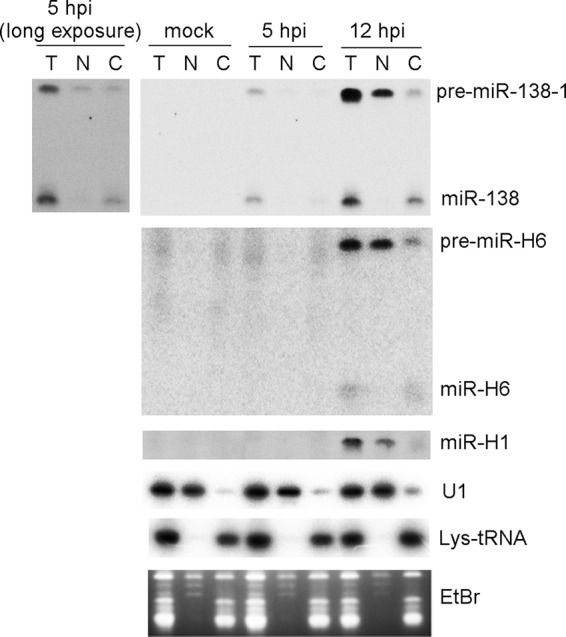
Nuclear export of pre-miRNA is blocked at late times during HSV-1 lytic infection. 293T cells were mock infected or infected with WTLyt138 virus (MOI = 5) for 5 or 12 h, at which times cells were harvested either for direct RNA purification (total) or for separation into nuclear and cytoplasmic fractions prior to RNA purification. The resulting RNAs were subjected to Northern blot hybridization for analysis of the RNAs indicated to the right of the panels. Mock infection and times postinfection (pi) are indicated at the top of the panel. The fractions are identified above the images. T, total RNA; N, nuclear fraction; C, cytoplasmic fraction. Mature miR-H1 was not detected in this experiment. The gel stained with ethidium bromide at ∼80 to ∼110 bases is shown in the bottom panel. An image from a longer exposure of the miR-138 signals is shown to the left of the main panel for miR-138. This experiment was performed twice. Additionally, similar results were obtained for miR-H6 in KOS-infected or KOS1.1-infected cells (Fig. 7E; see also Fig. S4).
Nuclear export of pre-miR-H6 is blocked at late times during HSV-1 lytic infection. 293T cells were mock infected or infected with HSV-1 strain KOS for 5 or 12 h, and cells were harvested at both times for either direct RNA purification (total) or for separation into nuclear and cytoplasmic fractions prior to RNA purification. The resulting RNAs were subjected to Northern blot hybridization, probing for the RNAs indicated to the right of the panels. Mock infection times and times postinfection are indicated at the top of the panel. The fractions are indicated above the images. T, total RNA; N, nuclear fraction; C, cytoplasmic fraction. This experiment was performed multiple times. Similar results were obtained in an experiment using WTLyt138 virus (Fig. 6). Download FIG S4, PDF file, 0.3 MB (295.7KB, pdf) .
Copyright © 2019 Pan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
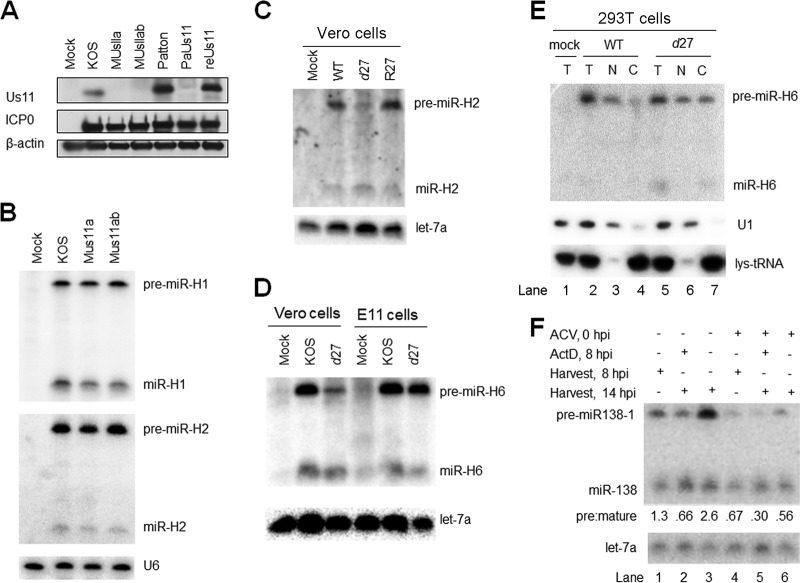
ICP27 and viral DNA synthesis but not Us11 are necessary for blocking pre-miRNA processing.
Hypothesizing that viral gene products might be involved in impeding miRNA biogenesis, we first tested the idea that Us11, an RNA binding protein with affinity for double-stranded RNA and even higher affinity for imperfect stem-loops (29, 30), might sequester pre-miRNAs, especially as Us11 has been suggested to block RNA silencing (31). However, pAUs11 mutant virus derived from strain Patton (32), which is Us11-defective due to a polyadenylation signal 5′ to the Us11 start codon, showed pre/mature ratios for miR-H1 and miR-H2 similar to those of the corresponding rescued virus (data not shown). Because we detected a weak immunoreactive band that comigrated with strain Patton Us11 in pAUs11-infected cells on a Western blot (Fig. 7A), we wondered whether leaky expression might be sufficient to impede miRNA biogenesis. We therefore constructed two additional Us11 mutant viruses on the basis of our KOS-derived WT-BAC virus (33). One virus, named MUs11a, removes an A from the Us11 start codon, which should result in translation starting from an out-of-frame AUG. The other virus, named MUs11ab, further replaces the second and third in-frame ATG with TGA stop codons in the MUs11a virus. Ablation of Us11 immunoreactivity comigrating with KOS Us11 was confirmed for both viruses (Fig. 7A). Both viruses exhibited levels of pre-miR-H1 and pre-miR-H2 and mature miR-H1 and miR-H2 similar to those seen with the WT (Fig. 7B), suggesting that Us11 is dispensable for regulation of miRNA biogenesis.
FIG 7.
Impairment of miRNA biogenesis is dependent on ICP27 and viral DNA synthesis but not on Us11. (A) Us11 expression from WT and mutant viruses. 293T cells were infected with the viruses indicated at the top of the panel (MOI = 5) and harvested at 16 hpi for Western blot analysis. The proteins analyzed are identified to the left of the panel. This experiment was performed three times. (B) Us11 deletion does not affect viral miRNA biogenesis. 293T cells were infected (MOI = 5) with the viruses indicated at the top of the panel. At 16 hpi, cells were harvested and analyzed by Northern blot hybridization for miR-H1 and miR-H2 expression. This experiment was performed several times. (C) Effects of ICP27 deletion on miR-H2 biogenesis. Vero cells were mock infected or infected (MOI = 20) for 24 h with the viruses indicated at the top of the panel. Pre-miR-H2 and miR-H2 expression (top panel) and let-7a expression (bottom) were analyzed by Northern blot hybridization. This experiment was performed twice. (D) Effects of ICP27 deletion on miR-H6 biogenesis in Vero and E11 cells. Vero cells, or E11 cells, which complement ICP27 expression, were infected with the viruses indicated at the top of the panel (MOI = 20) for 24 h, when the cells were analyzed by Northern blot hybridization for miR-H6. Expression of let-7a as a loading control is shown at the bottom. This experiment was performed twice. (E) Relief of the block of nuclear export in the absence of ICP27. 293T cells were mock infected (lane 1) or infected with the viruses indicated at the top of the panel. At 12 hpi, total RNA was isolated from infected cells (T; lanes 1, 2, and 5) or from nuclear (N; lanes 3 and 6) or cytoplasmic (C; lanes 4 and 7) fractions and analyzed as described for Fig. 6 for miR-H6, U1, and lys-tRNA (as indicated to the right of the panel). This experiment was performed twice. (F) Effects of ACV and ActD on miRNA biogenesis. 293T cells were infected (MOI = 5) with WT-BAC (lane 1) as a negative control for miR-138 expression or with WTLyt138 (lanes 2 to 7), as indicated in the top two lines of the panel. Plus signs (+) on the next two lines indicate treatment with ACV at the time of infection (lanes 5 to 7) or with ActD at 8 hpi (lanes 3 and 6), respectively. ACV was added to block viral DNA synthesis, and ActD was added to block RNA synthesis to help assess the stability of pre-miR-138 and mature miR-138. Plus signs on the next two lines indicate whether infected cells were harvested for RNA isolation at 8 hpi (lanes 2 and 5) or 14 hpi (lanes 1, 3, 4, 6, and 7). The image below shows the results of Northern analysis of expression of pre-miR-138-1 and miR-138, as indicated to the left. Pre/mature ratios calculated following band quantification are displayed below the image. let-7a levels as loading controls are shown at the bottom. This experiment was performed twice.
Another candidate for impeding miRNA biogenesis is ICP27, which plays a role in mRNA nuclear export (reviewed in reference 34). A homolog of ICP27 has been reported to block RNA silencing in plants (35). To test its involvement in miRNA biogenesis, we utilized an ICP27-null virus, d27 derived from HSV-1 strain KOS1.1 (36). Following infection of Vero cells, d27 expressed similar or possibly greater amounts of mature miR-H2 but substantially lower amounts of pre-miR-H2 than did both its parental WT virus and the rescued derivative, R27 (Fig. 7C). In another experiment, d27 expressed amounts of mature miR-H6 similar to those seen with the WT virus but much less pre-miR-H6 than the WT virus in infected Vero cells, and yet the d27 and WT viruses expressed similar amounts of pre-miR-H6 and mature miR-H6 in E11 cells, which ectopically express ICP4 and ICP27 (37) (Fig. 7D). In a third experiment, total RNA or RNA from nuclear and cytoplasmic fractions was isolated from WT and d27-infected 293T cells at 12 hpi. At that time point, similar to the results seen at the 12-hpi point (Fig. 6), WT virus expressed readily detectable pre-miR-H6 but much less mature miR-H6 (Fig. 7E, lane 2). The pre-miR-H6 was primarily found in the nuclear fraction (lane 3), with substantially less seen in the cytoplasmic fraction, and that small amount could be accounted for by nuclear contamination as evidenced by detection of U1 snRNA (lane 4). In contrast, the ratio of pre-miR-H6 to mature miR-H6 was lower in d27-infected cells than in WT-infected cells at 12 hpi (compare lanes 5 and 1), and the levels of pre-miR-H6 were similar in the nuclear and cytoplasmic fractions, which were cleanly separated based on the U1 and lys-tRNA markers. Taken together, these results show that ICP27 contributes to high pre/mature ratios and to retention of pre-miRNA in the nucleus.
Finally, we tested whether the impairment in miRNA biogenesis is dependent on viral DNA synthesis. 293T cells were infected with WTLyt138 in the absence or presence of acyclovir (ACV), an inhibitor of HSV DNA synthesis. At 8 hpi, infected cells were either mock treated or treated with ActD, and RNA was harvested immediately or at 14 hpi. In the absence of ACV and ActD, the miR-138 pre/mature ratios increased between 8 and 14 hpi (Fig. 7F, lanes 2 and 4), similarly to what we observed in Fig. 5D. Adding ActD in the absence of ACV at 8 hpi resulted in little change in pre-miR138-1 and mature miR-138 levels between 8 hpi and 14 hpi (lanes 2 and 3), consistent with the data in Fig. 4D indicating inefficient conversion of pre-miRNA to mature miRNA. However, in the presence of ACV, at both 8 and 14 hpi, the levels of mature miR-138 were similar to those seen in the absence of ACV at the same time points, but the levels of pre-miR-138-1 were considerably lower (compare lanes 5 and 7 with lanes 2 and 4), similarly to what we observed in the absence of ICP27 (Fig. 7C to E). Addition of ActD to ACV-treated infected cells at 8 hpi resulted in a decrease in pre-miR-138 and an increase in mature miR-138 (compare lanes 5 and 6), indicating conversion of pre-miRNA to mature miRNA. Thus, high pre/mature ratios are dependent on both ICP27 and viral DNA synthesis but not on Us11.
DISCUSSION
By characterizing HSV-1 pre-miRNA and miRNA expression in transduced and infected cells and in mouse ganglia, we found that although these miRNAs differed in inherent biogenesis efficiencies, conversion of pre-miRNA to miRNA was considerably less efficient during lytic infection than during latent infection or following lentiviral transduction, regardless of the miRNAs or cell types tested. Much if not all of this impairment of miRNA biogenesis was due to a block in pre-miRNA export from the nucleus to the cytoplasm. High pre/mature ratios occurred late in infection and were dependent on ICP27 and viral DNA synthesis but not on Us11. Below, we discuss these results and their implications for HSV-1 infection.
Regulation of miRNA biogenesis during HSV-1 lytic infection and variability in intrinsic biogenesis efficiency both contribute to miRNA expression.
Our Northern results show that all HSV-1 miRNAs tested exhibited high pre/mature ratios during lytic infection of multiple cell lines. Consistent with this, most exhibited high pre/mature ratios in ganglia at 3 dpi, arguing for a regulatory mechanism that applies to both acute ganglionic infection and lytic infection of cultured cells. During latency, pre/mature ratios were considerably reduced, arguing that miRNA biogenesis became more efficient, presumably due to reduced expression of the viral function(s) that impaired miRNA biogenesis. This mechanism then likely contributed to high expression of these miRNAs during latency.
Based on our results from lentivirus-transduced cells, the order of intrinsic efficiencies of conversion of pre-miRNA to miRNA is miR-H4 > miR-H2 > miR-H3, consistent with the relative pre/mature ratios for these miRNAs in infected cells and ganglia and suggesting that intrinsic biogenesis efficiencies contribute to determining expression levels. The finding of different intrinsic biogenesis efficiencies explains the inability to detect mature miR-H3 during lytic or latent infection by Northern analysis and the inability to detect pre-miR-H4 in infected ganglia. Variability in intrinsic biogenesis efficiency may reflect sequence-specific differences in pre-miRNA export and/or Dicer cleavage (38, 39). Regardless, for each miRNA tested, including the host miR-138, pre-miRNA conversion to miRNA was more efficient in lentivirus-transduced cells than in HSV-1 lytically infected cells, consistent with impairment of miRNA biogenesis during lytic infection.
Mechanism of impairment of miRNA biogenesis during lytic infection.
miRNA biogenesis during lytic infection of cells is temporally regulated. When detectable at early times of infection (e.g., in the case of ectopically expressed miR-138), pre-miRNAs convert efficiently to miRNAs, but the level of conversion is reduced later in infection. Moreover, pre-miRNAs exhibit short half-lives in the presence of ActD at 2 hpi but are much more stable at 8 hpi. Thus, impairment of miRNA biogenesis is a late infection event. Consistent with this, ACV treatment resulted in both reduced pre/mature ratios and reduced stability of pre-miRNA in the presence of ActD. Similarly, our finding that pre-miRNAs were depleted in the cytoplasm and enriched in the nucleus at 12 hpi, but not at 5 hpi, indicates that inhibition of nuclear export of pre-miRNAs occurs late. Thus, the results strongly suggest that inhibition of nuclear export of pre-miRNAs explains the impairment of miRNA biogenesis late during lytic infection. However, we cannot exclude the possibility of effects on other steps. Blocking nuclear export of pre-miRNAs is likely not unique to HSV-1 infection. Baculovirus encodes a miRNA that was reported to target the Exportin-5 cofactor Ran (40). Accumulation of pre-miR-31 in the nucleus with undetectable mature miR-31 has been reported in certain cancer cell lines (41), while activation of the oncogenic protein kinase extracellular signal-regulated kinase (ERK) has been reported to downregulate nuclear export of pre-miRNAs in hepatocellular carcinoma cells (42). Understanding how HSV blocks nuclear export of pre-miRNAs may reveal mechanisms that regulate miRNA biogenesis in other viral and cellular systems.
Deletion of ICP27 also greatly reduced pre/mature ratios for HSV-1 miRNAs. Interestingly, this deletion reduced the expression of the pre-miRNAs with no reduction or possibly an increase in the levels of the mature miRNAs, which we suggest was due to these pre-miRNAs being late viral gene products (12) and thus likely to be dependent on ICP27 for their full expression. Nevertheless, the simplest interpretation is that ICP27, one of whose many functions (34) is to promote export of certain mRNAs (43–45), is required for impairment of miRNA biogenesis. Assuming that this is correct, the issue becomes whether ICP27 itself acts to impair miRNA biogenesis or whether ICP27 is required for synthesis of one or more viral gene products that do so. Supporting the former possibility, the turkey herpesvirus (HVT) homolog of ICP27 has been reported to be sufficient for blocking RNA silencing in tobacco (35), and it has been speculated that ICP27 may block nuclear export of certain RNAs (46). Supporting the latter possibility are the immediate early expression of ICP27 versus the late impairment of miRNA biogenesis and the similar phenotypes of ACV treatment and ICP27 deletion, especially given that most viral late and certain DNA replication proteins require ICP27 for their efficient synthesis (47). A third possibility is that HSV-1 induces expression of a host factor that impairs miRNA biogenesis in an ICP27-dependent fashion. Experiments are under way to test these alternatives.
Implications for HSV-1 infection.
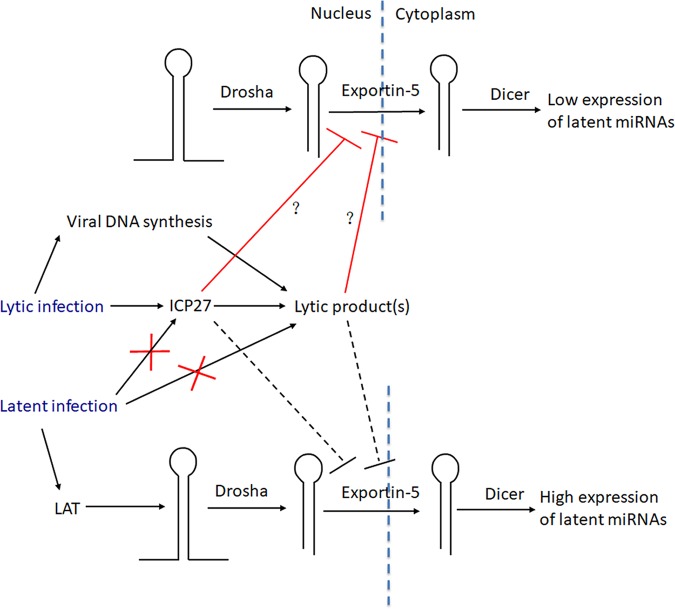
Our results lead to a model (Fig. 8) in which HSV-1 lytic infection blocks pre-miRNA conversion to miRNAs at the stage of nuclear export whereas lytic gene products responsible for this block are depleted during latent infection, allowing more efficient biogenesis. Certain HSV miRNAs that are expressed abundantly during latency have been shown to be able to repress expression of important lytic proteins such as ICP0, ICP4, and ICP34.5 in transfection experiments (3, 9, 10, 13–15). miRNAs that are abundant during latency might also inhibit the expression of host proteins that would otherwise promote lytic rather than latent infection. Thus, latent HSV miRNAs have been hypothesized to promote latency. Although some in vivo mutational studies of HSV-1 and HSV-2 miRNAs have yielded negative results (15, 25), interpretation of these results is complicated by the possibility of functional redundancy and the limitations of using small animal models to recapitulate latency in humans using small-animal models. Blocking miRNA biogenesis during lytic infection may serve to limit the extent to which expression of their targets is repressed, allowing less-restricted viral replication. This proposal is supported by a study that showed that HSV-1 infection suppresses RNA-induced silencing and that knockdown of Ago2 expression increases HSV-1 replication (48). During latent infection, when repression of lytic gene expression (or certain host factors) is beneficial to the virus, removal of the block to miRNA biogenesis combined with high transcription of the LAT locus permits strong expression of latent miRNAs (Fig. 8).
FIG 8.
Cartoon of proposed mechanism of regulation of miRNA biogenesis during HSV-1 infection. The cartoon is divided into a nuclear portion to the left and a cytoplasmic portion to the right, with vertical dotted lines representing the nuclear membrane. The upper and lower parts of the cartoon illustrate events occurring during lytic infection and latent infection, respectively. Pre-miRNA stem-loop structures are represented by hairpins. Lines terminating with arrowheads point from causes to results of events, with important factors shown above the arrows and with each red X indicating a loss of expression of important factors. Red lines terminating with perpendicular lines indicate events being blocked by ICP27 and/or lytic products dependent upon ICP27 for their synthesis, with the dashed line indicating a loss of the block.
In plants and invertebrates, viruses rely on virus-encoded suppressors of RNA interference (RNAi) to combat an RNAi-mediated antiviral response, although the functional relevance of this suppressive activity in mammals remains to be established (49). Mammalian viruses, including some herpesviruses, have displayed various ways of regulating miRNA biogenesis (reviewed in references 19 and 50). However, we are unaware of any previous examples of regulation of pre-miRNA nuclear export by mammalian viruses. Effects of viral infection on expression of regulators of miRNA biogenesis have been observed with gammaherpesviruses (see, e.g., references 51 and 52), and there is evidence that levels of certain mature viral miRNAs are regulated posttranscriptionally (see, e.g., references 53 and 54). Specific downregulation of a host miRNA by overexpression of a complementary target has been reported previously for both betaherpesviruses and gammaherpesviruses (55–58). However, although the high pre/mature ratios of most viral miRNAs that we document here can also be found during lytic infection with HSV-2 and pseudorabies (9, 10, 24, 25, 59), they have not been generally observed during the lytic cycles of the avian alphaherpesvirus, Marek’s disease virus, betaherpesviruses, or gammaherpesviruses (60–66). Therefore, general impairment of conversion of pre-miRNA to mature miRNA appears to be found mainly among mammalian alphaherpesviruses. We speculate that this might relate to their neurotropism. Regardless, our findings identify a novel mechanism of differential regulation of miRNA expression that may ensure optimal outcomes for lytic and latent infection.
MATERIALS AND METHODS
Cells and viruses.
Vero, 293T, and Neuro-2A cells were maintained as previously described (28). Vero and Neuro-2A cells were obtained from ATCC and used following less than 6 months of passage. 293T cells were authenticated by small-tandem-repeat profiling at the Dana-Farber Cancer Institute Molecular Diagnostics Laboratory in December 2018. Viruses were propagated in Vero cells and titrated by plaque assays following standard protocols (67). WTLyt138, MUs11a, and MUs11b viruses were constructed based on a BAC-containing strain, KOS (33), by two-step red recombination (68). To construct WTLyt138 virus, the following human pre-miR-138-1 and flanking sequences were synthesized as a gBlocks gene fragment (Integrated DNA Technologies): CTTAGAATTCGGATCATCAATCATCTGCCTTCCCAGGACCACCCAGCAGTGTTGCACGTAGAGCAGAGGTGCAGTGCTGCACGTACAGCAGAGGCGACCCCTTCTCCATACTTCAGAGACCTCTAGCATCGTGTTGTGGGACAGCTGGTGTTGTGAATCAGGCCGTTGCCAATCAGAGAACGGCTACTTCACAACACCAGGGCCACACTGCACTGCAAGCAGCGAGCAGCAAGCAACAAGCATCCAGACACCTGAGCACCAGGCTGCCAGCAAGAGCACCTGCCTGTGAGGAAACCTCCCAGCCCACCCCATACCCCACCCCATTATCCCACCCTACAAGCTTGGTA. The sequences were cloned between the EcoRI and HindIII sites of FLAG-HA-pcDNA3.1(-) plasmid (AddGene). A kanamycin resistance gene (Kan) flanked by one I-SceI site was then cloned into the EcoRI site of the resulting plasmid using the following two primers: primer CTTAGAATTCGGATCATCAATCATCTGCCTTCCCAGGACCACCCAGCAGTGTTGCACGTATAGGGATAACAGGGTAATCGATTT and primer CTTAGAATTCGCCAGTGTTACAACCAATTAACC. The resulting plasmid was used as a template to amplify a cassette to be recombined with the HSV-1 genome using the following two primers: primer AATCCGGTAACCCGTTGAGTCCCGGGTACGACCATCACCCGAGTCTCTGGAATTCGGATCATCAATCATCTGCC and primer GGGTTGGGTCTGGCTCATCTCGAGAGACACGGGGGGGAACCACCCTCCGCGTAGGGTGGGATAATGGGGTGG. The subsequent recombination of this cassette with the viral genome and removal of Kan were conducted as described previously (68). The two-step red recombination protocol was also used to construct MUs11a BAC using primer TACGACCATCACCCGAGTCTCTGGGCGGAGGGTGGTTCCCCCCCGTGTCTCTCGAGTGAGCCAGACCCAAcCCCCGTAGGGATAACAGGGTAATCGATTT and primer TAAGTAAACATCTGGGTCGCCCGGCCCAACTGGGGCCGGGGGTTGGGTCTGGCTCACTCGAGAGACACGGGGGGGAGCCAGTGTTACAACCAATTAACC as well as to construct MUs11b based on MUs11a using primer TGTTTACTTAAAAGGCGTGCCGTCCGCCGGCTGACACCCCAGAGGTGTTCACGCACCTCGAGGACACCCGCGCTGATAGGGATAACAGGGTAATCGATTT and primer TCATTATCACCCCGTTGCGGGGGTCCGgAGATTCAGCGCGGGTGTCCTCGAGGTGCGTGAACACCTCTGGGGTGTCAGCCAGTGTTACAACCAATTAACC.
Viruses were produced by transfecting BAC DNAs into Vero cells using Lipofectamine 3000 following the instructions of the manufacturer (Thermo Fisher). Cells were infected by adding virus at the indicated MOI, incubating for 1 h at 37°C with gentle shaking every 15 min for adsorption, and replacing the supernatant with fresh media and incubating at 37°C for the indicated times.
Northern blot hybridization.
RNAs of <200 nucleotides were purified from cultured cells or pooled mouse ganglia using a miRNeasy minikit (Qiagen) following the manufacturer’s instructions and were resolved on Novex 15% Tris-borate-EDTA-urea polyacrylamide gels (Thermo Fisher). We used radioactive and nonradioactive methods interchangeably for Northern blot hybridization. The nonradioactive method followed a previously described protocol (15). LNA probes were synthesized and 3′ end labeled with digoxigenin by Exiqon. The radioactive method followed the same protocol before hybridization. During the prehybridization step, 10 pmol of probe was incubated with 1 μl of T4 polynucleotide kinase (NEB), 2.5 μl of the supplied buffer, and 1 to 5 μl of [γ-32P]ATP (Perkin Elmer) in a total volume of 25 μl for 1 h at 37°C before this mixture was added into the hybridization tube. The subsequent hybridization and washing conditions were the same as those used with the nonradioactive method. After the last wash, the membrane was exposed to a phosphor screen and visualized by a phosphorimager. Band intensities were quantified by Quantity One 1-D Analysis Software (Bio-Rad). The probe sequences and optimized hybridization and washing temperatures for different RNAs are listed in Table 1.
TABLE 1.
Sequences of Northern blot probes
| Probe designation |
Probe sequencea | Optimized hybridization and washing temp (°C)b |
|---|---|---|
| miR-H1 | TCCACTTC+CC+G+TC+C+TTCCATC | 60 |
| miR-H2 | AGTCGC+AC+TCG+T+CCCTGGCTCAGG | 60 |
| miR-H3 | GTCCCA+A+C+C+G+C+A+C+A+GTCCCAG | 65 |
| miR-H4 | ACTAGC+G+AG+TT+AGA+C+AGGCAAG | 55 |
| miR-H5 | AGGG+TT+T+GG+ATC+T+CTGAC | 60 |
| miR-H6 | GGGA+TG+GA+AG+GA+CGGG+AAGTG | 55 |
| miR-138 | CGGCCTG+A+TT+C+A+CA+ACACCAGCT | 60 |
| let-7a | AACTAT+A+CA+A+CCT+A+CTACCTCA | 60 |
| U6 | CGAAT+TTG+CGT+GT+CAT+CCTTGC | 65 |
| U1 | CGAGTTTCCCACATTTGGGG | 37 |
| lys-tRNA | CTGATGCTCTACCGACTGAGCTATCCGGGC | 37 |
The probes are labeled with digoxigenin at the 3’ end, for nonradioactive detection. +N, locked nucleic acid-modified nucleotide.
The washing temperature is typically the same as hybridization temperature, but during the last two washes, raising the temperature by 5°C helps further remove nonspecific bands.
Animal procedures.
The animal housing and experimental procedures were approved by the Institutional Animal Care and Use Committee of Harvard Medical School in accordance with federal guidelines. Male CD-1 mice (Charles River Laboratories) (7 weeks old) were infected with HSV-1 strain KOS as described previously (15). At the indicated times, trigeminal ganglia were removed and placed on dry ice before being stored at −80°C. For reactivation, trigeminal ganglia removed from mice at 30 dpi were held in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum on ice during the dissection procedure and were then incubated at 37°C for 1 to 3 days before the ganglia were transferred to Eppendorf tubes. Sixteen ganglia were pooled for each condition before RNA purification.
Construction of lentivirus-transduced cell lines.
The synthesized gBlocks gene fragment described above was used as a miR-138-expressing sequence. miR-H2-expressing and miR-H3/H4-expressing sequences were amplified from HSV-1 WT BAC using primers TGACCTCGAGTTTTCCTGGCCCGACCCGCGCCTCTT and CTGTGAATTCGACCCCATAGTGATCAGCGACTCC for miR-H2 and primers TGACCTCGAGGACGCGGACTCGGGAACGTGGA and CTGTGAATTCTCGCGCGCGGCCCTTTAAAGGCG for miR-H3/H4. Each miRNA-expressing sequence was inserted between the XhoI and EcoRI restriction sites of pTRIPZ (69) (a generous gift from Bryan Cullen). A 4-μg amount of each of the resulting plasmids was transfected together with 7.1 μg of psPAX2 plasmid (Addgene) and 3.9 μg of pMD2.G plasmid (Addgene) into 293T cells in a 100-mm-diameter plate using Lipofectamine 3000 following the instructions of the manufacturer (Thermo Fisher). Lentiviruses were harvested from the supernatants 3 days posttransfection and added to 50% confluent 293T or Neuro-2a cells in the presence of 8 μg/ml of hexadimethrine bromide (Sigma). Two days later, the supernatant was removed and replaced with fresh medium containing 1 μg/ml of puromycin. Surviving cells were expanded in the presence of puromycin until the cells were stably maintained.
Western blotting.
Western blotting was performed as described previously (57). The following primary antibodies (manufacturer and catalog number or reference number) were used at the indicated dilutions: ICP0 antibody (Abcam, ab6513), 1:5,000; ICP27 antibody (Virusys, 1113), 1:5,000; β-actin antibody (Sigma, A5441), 1:10,000; Drosha antibody (Abcam, ab12286), 1:1,000; Dicer antibody (Santa Cruz, H212), 1:200; Exportin 5 antibody (Santa Cruz, H-300), 1:200; Us11 antibody (70), 1:1,000.
Cell fractionation.
Cell fractionation was performed using a tissue grinder (Wheaton) (1 ml) as previously described (71). RNA was purified from total, cytoplasmic, and nuclear fractions using a mirVana miRNA isolation kit following the instructions of the manufacturer (Ambion).
ACKNOWLEDGMENTS
We thank Richard Gregory and Eain Murphy for valuable advice, Ian Mohr for generous provision of WT, mutant, and rescued viruses, Bryan Cullen for generous provision of pTRIPZ plasmid, and Margarita Krivitski and Soo-Mi Lee for preliminary studies and helpful discussions.
This work was supported by grants from the National Key R&D Program of China (2017YFC1200204 to D.P.), the National Natural Science Foundation of China (81671993 to D.P.), and Zhejiang Provincial Natural Science Foundation of China (LR18H190001 to D.P.); the National Institutes of Health (F31 AI129207 to M.E.M. and P01 AI098681 to D.M.C. and D.M.K.); and the Croatian Science Foundation (8790 to I.J.).
Footnotes
Citation Pan D, Li G, Morris-Love J, Qi S, Feng L, Mertens ME, Jurak I, Knipe DM, Coen DM. 2019. Herpes simplex virus 1 lytic infection blocks microRNA (miRNA) biogenesis at the stage of nuclear export of pre-miRNAs. mBio 10:e02856-18. https://doi.org/10.1128/mBio.02856-18.
Contributor Information
Xiang-Jin Meng, Virginia Polytechnic Institute and State University.
Rozanne Sandri-Goldin, University of California, Irvine.
Nancy Raab-Traub, University of North Carolina at Chapel Hill.
REFERENCES
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex virus, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racianello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol 80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol 84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Z, Liu X, Chen X, Zhou X, Du T, Roizman B, Zhou G. 2016. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc Natl Acad Sci U S A 113:E894–E901. doi: 10.1073/pnas.1525674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu W, Guo Z, Zhang X, Guo L, Liu L, Liao Y, Wang J, Wang L, Li Q. 2013. A microRNA encoded by HSV-1 inhibits a cellular transcriptional repressor of viral immediate early and early genes. Sci China Life Sci 56:373–383. doi: 10.1007/s11427-013-4458-4. [DOI] [PubMed] [Google Scholar]
- 7.Munson DJ, Burch AD. 2012. A novel miRNA produced during lytic HSV-1 infection is important for efficient replication in tissue culture. Arch Virol 157:1677–1688. doi: 10.1007/s00705-012-1345-4. [DOI] [PubMed] [Google Scholar]
- 8.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. 2009. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol 83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A 105:10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang S, Patel A, Krause PR. 2009. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol 83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang S, Bertke AS, Patel A, Margolis TP, Krause PR. 2011. Herpes simplex virus 2 microRNA miR-H6 is a novel latency-associated transcript-associated microRNA, but reduction of its expression does not influence the establishment of viral latency or the recurrence phenotype. J Virol 85:4501–4509. doi: 10.1128/JVI.01997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer MF, Jurak I, Pesola JM, Boissel S, Knipe DM, Coen DM. 2011. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology 417:239–247. doi: 10.1016/j.virol.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores O, Nakayama S, Whisnant AW, Javanbakht H, Cullen BR, Bloom DC. 2013. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J Virol 87:6589–6603. doi: 10.1128/JVI.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Brown D, Osorio N, Hsiang C, Li L, Chan L, BenMohamed L, Wechsler SL. 2015. A herpes simplex virus type 1 mutant disrupted for microRNA H2 with increased neurovirulence and rate of reactivation. J Neurovirol 21:199–209. doi: 10.1007/s13365-015-0319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan D, Pesola JM, Li G, McCarron S, Coen DM. 2017. Mutations inactivating herpes simplex virus-1 microRNA miR-H2 do not detectably increase ICP0 gene expression in infected cultured cells or mouse trigeminal ganglia. J Virol 91:e02001-16. doi: 10.1128/JVI.02001-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naqvi AR, Seal A, Shango J, Brambila MF, Martinez G, Chapa G, Hasan S, Yadavalli T, Jaishankar D, Shukla D, Nares S. 2018. Herpesvirus-encoded microRNAs detected in human gingiva alter host cell transcriptome and regulate viral infection. Biochim Biophys Acta Gene Regul Mech 1861:497–508. doi: 10.1016/j.bbagrm.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurak I, Silverstein LB, Sharma M, Coen DM. 2012. Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity. J Virol 86:10093–10102. doi: 10.1128/JVI.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enk J, Levi A, Weisblum Y, Yamin R, Charpak-Amikam Y, Wolf DG, Mandelboim O. 2016. HSV1 microRNA modulation of GPI anchoring and downstream immune evasion. Cell Rep 17:949–956. doi: 10.1016/j.celrep.2016.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. 2018. Metazoan microRNAs. Cell 173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 21.Jurak I, Hackenberg M, Kim JY, Pesola JM, Everett RD, Preston CM, Wilson AC, Coen DM. 2014. Expression of herpes simplex virus 1 microRNAs in cell culture models of quiescent and latent infection. J Virol 88:2337–2339. doi: 10.1128/JVI.03486-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du T, Han Z, Zhou G, Zhou G, Roizman B. 2015. Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation. Proc Natl Acad Sci U S A 112:E49–E55. doi: 10.1073/pnas.1422657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Held K, Junker A, Dornmair K, Meinl E, Sinicina I, Brandt T, Theil D, Derfuss T. 2011. Expression of herpes simplex virus 1-encoded microRNAs in human trigeminal ganglia and their relation to local T-cell infiltrates. J Virol 85:9680–9685. doi: 10.1128/JVI.00874-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang S, Bosch-Marce M, Patel A, Margolis TP, Krause PR. 2015. Characterization of herpes simplex virus 2 primary microRNA transcript regulation. J Virol 89:4837–4848. doi: 10.1128/JVI.03135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura Y, Bosch-Marce M, Tang S, Patel A, Krause PR. 2018. Herpes simplex virus 2 latency-associated transcript (LAT) region mutations do not identify a role for LAT-associated microRNAs in viral reactivation in the guinea pig genital model. J Virol 92:e00642-18. doi: 10.1128/JVI.00642-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du T, Zhou G, Roizman B. 2011. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci U S A 108:18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivas HG, Schmaling SK, Gaglia MM. 2016. Shutoff of host gene expression in influenza A virus and herpesviruses: similar mechanisms and common themes. Viruses 8:102. doi: 10.3390/v8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM. 2014. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe 15:446–456. doi: 10.1016/j.chom.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roller RJ, Roizman B. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J Virol 64:3463–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryant KF, Cox JC, Wang H, Hogle JM, Ellington AD, Coen DM. 2005. Binding of herpes simplex virus-1 US11 to specific RNA sequences. Nucleic Acids Res 33:6090–6100. doi: 10.1093/nar/gki919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z. 2008. Silencing suppression by herpes simplex virus type 1. PhD thesis. The Ohio State University, Columbus, OH. [Google Scholar]
- 32.Mulvey M, Poppers J, Sternberg D, Mohr I. 2003. Regulation of eIF2alpha phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J Virol 77:10917–10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan D, Coen DM. 2012. Quantification and analysis of thymidine kinase expression from acyclovir-resistant G-string insertion and deletion mutants in herpes simplex virus-infected cells. J Virol 86:4518–4526. doi: 10.1128/JVI.06995-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandri-Goldin RM. 2011. The many roles of the highly interactive HSV protein ICP27, a key regulator of infection. Future Microbiol 6:1261–1277. doi: 10.2217/fmb.11.119. [DOI] [PubMed] [Google Scholar]
- 35.Jing XL, Fan MN, Jia G, Liu LW, Ma L, Zheng CC, Zhu XP, Liu HM, Wang XY. 2011. A multifunctional protein encoded by turkey herpesvirus suppresses RNA silencing in Nicotiana benthamiana. J Virol 85:12792–12803. doi: 10.1128/JVI.05565-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice SA, Knipe DM. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J Virol 64:1704–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samaniego LA, Neiderhiser L, DeLuca NA. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol 72:3307–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, Zhang X, Graves P, Zeng Y. 2012. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA 18:2083–2092. doi: 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng Y, Cullen BR. 2004. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res 32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh CP, Singh J, Nagaraju J. 2012. A baculovirus-encoded microRNA (miRNA) suppresses its host miRNA biogenesis by regulating the Exportin-5 cofactor Ran. J Virol 86:7867–7879. doi: 10.1128/JVI.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. 2008. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun HL, Cui R, Zhou J, Teng KY, Hsiao YH, Nakanishi K, Fassan M, Luo Z, Shi G, Tili E, Kutay H, Lovat F, Vicentini C, Huang HL, Wang SW, Kim T, Zanesi N, Jeon YJ, Lee TJ, Guh JH, Hung MC, Ghoshal K, Teng CM, Peng Y, Croce CM. 2016. ERK activation globally downregulates miRNAs through phosphorylating Exportin-5. Cancer Cell 30:723–736. doi: 10.1016/j.ccell.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen IH, Sciabica KS, Sandri-Goldin RM. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J Virol 76:12877–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koffa MD, Clements JB, Izaurralde E, Wadd S, Wilson SA, Mattaj IW, Kuersten S. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J 20:5769–5778. doi: 10.1093/emboj/20.20.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson A, Knipe DM, Coen DM. 2004. ICP27 selectively regulates the cytoplasmic localization of a subset of viral transcripts in herpes simplex virus type 1-infected cells. J Virol 78:23–32. doi: 10.1128/JVI.78.1.23-32.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuss SK, Mata MA, Zhang L, Fontoura BM. 2013. Nuclear imprisonment: viral strategies to arrest host mRNA nuclear export. Viruses 5:1824–1849. doi: 10.3390/v5071824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uprichard SL, Knipe DM. 1996. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J Virol 70:1969–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Z, Zhu Y, Bisaro DM, Parris DS. 2009. Herpes simplex virus type 1 suppresses RNA-induced gene silencing in mammalian cells. J Virol 83:6652–6663. doi: 10.1128/JVI.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cullen BR, Cherry S, tenOever BR. 2013. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe 14:374–378. doi: 10.1016/j.chom.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 50.Libri V, Miesen P, van Rij RP, Buck AH. 2013. Regulation of microRNA biogenesis and turnover by animals and their viruses. Cell Mol Life Sci 70:3525–3544. doi: 10.1007/s00018-012-1257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Happel C, Ramalingam D, Ziegelbauer JM. 2016. Virus-mediated alterations in miRNA factors and degradation of viral miRNAs by MCPIP1. PLoS Biol 14:e2000998. doi: 10.1371/journal.pbio.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin YT, Sullivan CS. 2011. Expanding the role of Drosha to the regulation of viral gene expression. Proc Natl Acad Sci U S A 108:11229–11234. doi: 10.1073/pnas.1105799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edwards RH, Marquitz AR, Raab-Traub N. 2008. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J Virol 82:9094–9106. doi: 10.1128/JVI.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amoroso R, Fitzsimmons L, Thomas WA, Kelly GL, Rowe M, Bell AI. 2011. Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation. J Virol 85:996–1010. doi: 10.1128/JVI.01528-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. 2012. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc Natl Acad Sci U S A 109:279–284. doi: 10.1073/pnas.1114204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cazalla D, Yario T, Steitz JA, Steitz J. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, Zimmer R, Trgovcich J, Koszinowski UH, Jonjic S, Pfeffer S, Dölken L. 2012. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog 8:e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, Ahn K. 2013. Selective degradation of host microRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 13:678–690. doi: 10.1016/j.chom.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 59.Wu YQ, Chen DJ, He HB, Chen DS, Chen LL, Chen HC, Liu ZF. 2012. Pseudorabies virus infected porcine epithelial cell line generates a diverse set of host microRNAs and a special cluster of viral microRNAs. PLoS One 7:e30988. doi: 10.1371/journal.pone.0030988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. 2007. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol 81:13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol 7:1684–1695. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 62.Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol 79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuddenham L, Jung JS, Chane-Woon-Ming B, Dölken L, Pfeffer S. 2012. Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B. J Virol 86:1638–1649. doi: 10.1128/JVI.05911-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Samols MA, Hu J, Skalsky RL, Renne R. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol 79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A 102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res 36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coen DM, Fleming HE Jr, Leslie LK, Retondo MJ. 1985. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J Virol 53:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 69.Gottwein E, Cullen BR. 2010. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol 84:5229–5237. doi: 10.1128/JVI.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bryant KF, Macari ER, Malik N, Boyce M, Yuan J, Coen DM. 2008. ICP34.5-dependent and -independent activities of salubrinal in herpes simplex virus-1 infected cells. Virology 379:197–204. doi: 10.1016/j.virol.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.ten Have S, Hodge K, Lamond AI. 2012. Dynamic proteomics: methodologies and analysis In Meroni G, Petrera F (ed), Functional genomics. IntechOpen, London, United Kingdom. doi: 10.5772/50786. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HSV-1 pre-miRNA and mature miRNA expression in trigeminal ganglia at different times after infection or after reactivation from latency. Mice were mock infected or infected on the cornea with 2 × 105 PFU of HSV-1 strain KOS per eye. At various days postinfection (dpi), trigeminal ganglia were collected. At 30 dpi, some ganglia were explanted and cultivated for various days postexplantation (dpe) before the ganglia were harvested for RNA. Sixteen ganglia were pooled for each condition, and small RNAs (<200 bases) were isolated and analyzed by Northern blot hybridization. RNAs purified from 293T cells that had been mock infected or infected for 18 h were run alongside (see last two lanes) the RNAs from ganglia to provide markers. The sources of RNAs and time points are indicated at the tops of each series (left or right) of Northern images. Positions of miRNA species are indicated to the right of the images. Ethidium bromide signals at ∼80 bases from the gels used for each series of Northern images are displayed at the bottom. An independent experiment is shown in Fig. 1. Download FIG S1, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2019 Pan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Stability of pre-miRNAs and miRNAs in HSV-1-infected cells. 293T cells were infected with HSV-1 strain KOS (MOI = 10). At 2 (left panel) or 8 (right panel) hpi, 1 μg/ml ActD was added, and cells were harvested at the times indicated at the top of the images. miR-H4 was analyzed by Northern blot hybridization. Results of experiments examining miR-H2 are shown in Fig. 4C and D. Download FIG S2, PDF file, 0.4 MB (421.3KB, pdf) .
Copyright © 2019 Pan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HSV-1 infection does not reduce expression of various proteins involved in miRNA biogenesis. 293T and Vero cells were infected at an MOI of 10 and Neuro-2a cells were infected at an MOI of 30 with HSV-1 strain KOS for the times indicated at the top of the panel. The proteins indicated to the left were analyzed by Western blotting. The experiment using Vero and 293T cells was performed multiple times. The experiment using Neuro-2a cells was performed once. Download FIG S3, PDF file, 0.2 MB (255.1KB, pdf) .
Copyright © 2019 Pan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nuclear export of pre-miR-H6 is blocked at late times during HSV-1 lytic infection. 293T cells were mock infected or infected with HSV-1 strain KOS for 5 or 12 h, and cells were harvested at both times for either direct RNA purification (total) or for separation into nuclear and cytoplasmic fractions prior to RNA purification. The resulting RNAs were subjected to Northern blot hybridization, probing for the RNAs indicated to the right of the panels. Mock infection times and times postinfection are indicated at the top of the panel. The fractions are indicated above the images. T, total RNA; N, nuclear fraction; C, cytoplasmic fraction. This experiment was performed multiple times. Similar results were obtained in an experiment using WTLyt138 virus (Fig. 6). Download FIG S4, PDF file, 0.3 MB (295.7KB, pdf) .
Copyright © 2019 Pan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.