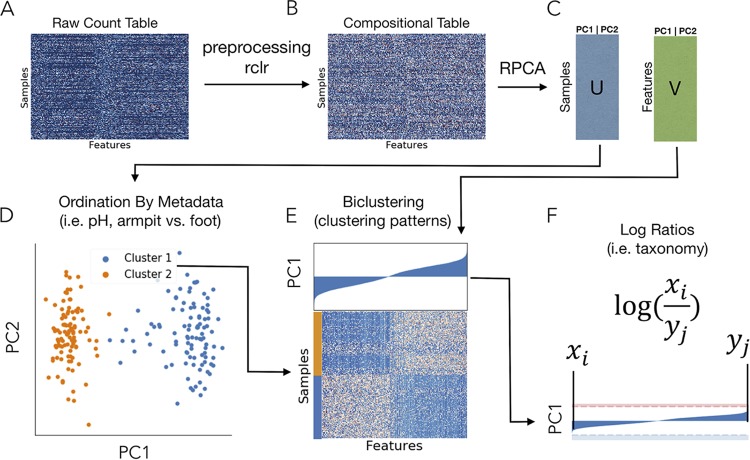
FIG 2.
A general overview of the workflow. (A) A sparse, raw sequencing count table with samples on the y axis and features (i.e., OTUs, genes) on the x axis. (B) The data are preprocessed by a robust centered log ratio transform (rclr) on only the known (nonzero) values. (C) Matrix completion with a robust principal-component analysis (RPCA) that operates on only the observed values in the table resolves a loading by samples and by features. These loadings can be directly used for ordination (D), biclustering (E), and the identification of important taxa driving clustering in both the previous plots (F).