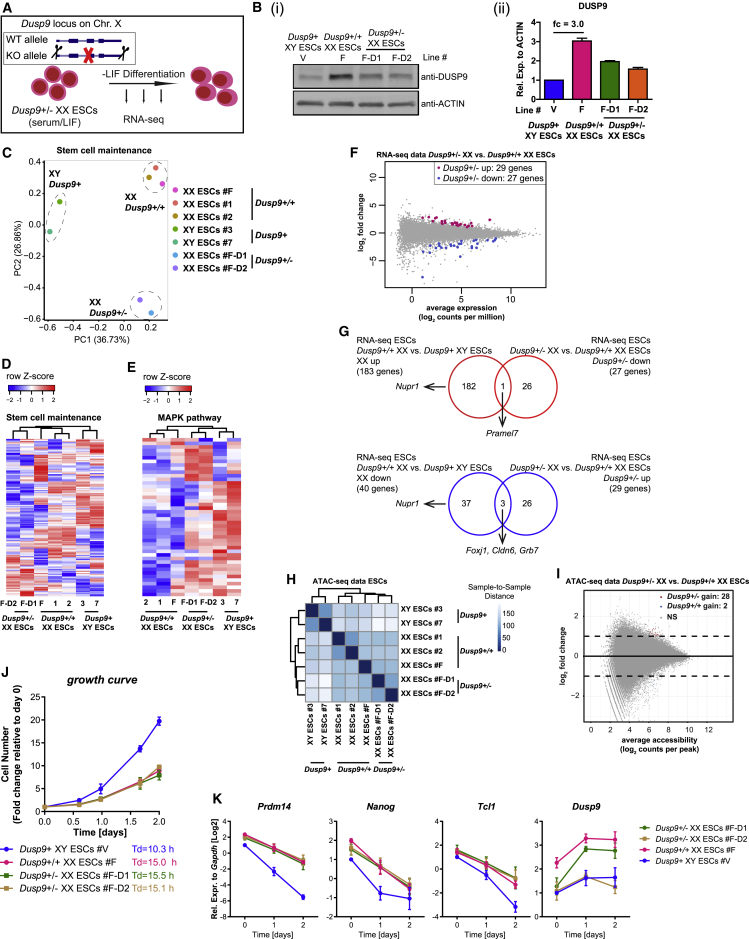
Figure 5.
The Open Chromatin Landscape, Growth, and Pluripotency Exit Delay of XX ESCs Are Maintained in the Presence of Male-like Global DNA Methylation
(A) Scheme of Dusp9 heterozygous deletion in XX ESCs followed by LIF withdrawal differentiation. KO, knockout; WT, wild-type.
(B) (i) Western blot analysis for DUSP9 in Dusp9+/− ESCs, Dusp9+/+ ESCs, and XY ESCs grown in S/L. (ii) Quantification of DUSP9 levels using actin as a loading control (n = 2).
(C) PCA of stem cell maintenance genes (RNA-seq data) for the Dusp9+/−, Dusp9+/+, and XY ESCs grown in S/L conditions.
(D) Unsupervised hierarchical clustering of stem cell maintenance genes for the Dusp9+/−, Dusp9+/+, and XY ESCs.
(E) As in (D) for MAPK pathway-related genes (defined in Schulz et al., 2014).
(F) DEG analysis, identifying clear differences between Dusp9+/+ XX and XY ESCs, but much less between Dusp9+/− XX and Dusp9+/+ XX ESCs.
(G) Overlap between the DEGs of Dusp9+/+ versus XY ESCs and Dusp9+/− versus Dusp9+/+ ESCs for upregulated genes (up) and downregulated genes (down). Dusp9 heterozygous deletion maintains a female-like transcriptome.
(H) ATAC-seq sample-to-sample distance heatmap in Dusp9+/+, Dusp9+/−, and XY ESCs. Dusp9+/− ESCs maintain a Dusp9+/+-like open chromatin landscape.
(I) Differential chromatin accessibility analysis between Dusp9 heterozygous mutant and wild-type XX ESCs. Log2 fold change (mutant/wild-type) in reads per accessible region are plotted against the mean reads per ATAC-seq peak. Dusp9 heterozygous mutants maintain a female-like open chromatin landscape.
(J) Growth curves and doubling times of Dusp9+/−, Dusp9+/+, and XY ESCs in S/L condition. Cells were counted at the indicated time points and presented as fold changes relative to day 0, averages (±SEM) of biological duplicates. Data from a representative experiment from at least 4 independent experiments.
(K) qRT-PCR for Prdm14, Nanog, Tcl1, and Dusp9 expression before and after LIF withdrawal (n = 3).
See also Figure S5.