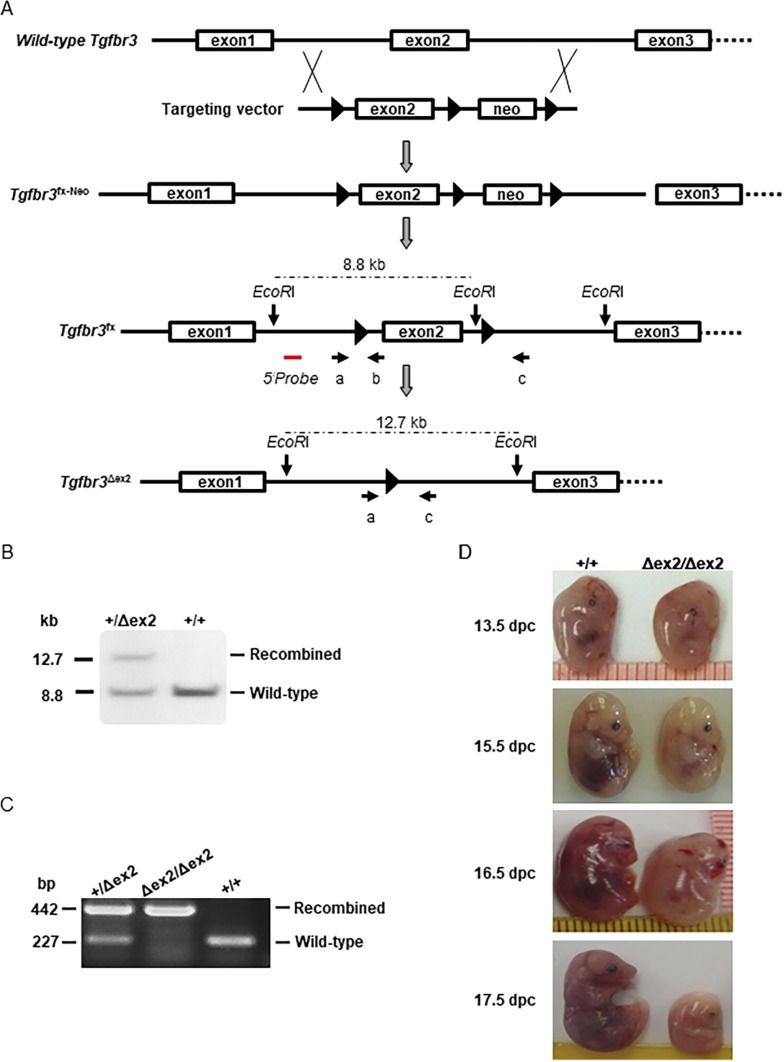
Figure 1.
Generation and validation of floxed Tgfbr3 mice. (A) Schematic representation shows the strategy used to generate the conditional Tgfbr3 allele. LoxP sites are pictured as black triangles, and exons are shown as boxes. The red line indicates the approximate position of the Southern blot probe used in (B). Primers used in PCR in (C) are shown as arrows and labeled a, b, and c. Note that exons, introns, the Southern probe, and primers are not drawn to scale. (B) Southern blot analysis of wild-type (+) and recombined (Δex2) Tgfbr3 alleles is shown. Genomic DNA was digested with EcoRI. The approximate locations of the EcoRI sites and restriction fragment sizes are indicated in (A). (C) Examples show PCR genotyping of heterozygous (Tgfbr3+/∆ex2), homozygous (Tgfbr3∆ex2/∆ex2), and wild-type (Tgfbr3+/+) mice. The wild-type allele was detected with primers a and b (note that the loxP site is absent in this allele but present in the floxed allele). The recombined allele was detected with primers a and c. (D) Examples show wild-type (+/+) and homozygous knockout (∆ex2/∆ex2) embryos at the indicated ages. dpc, day post coitus; neo, neomycin selection cassette.