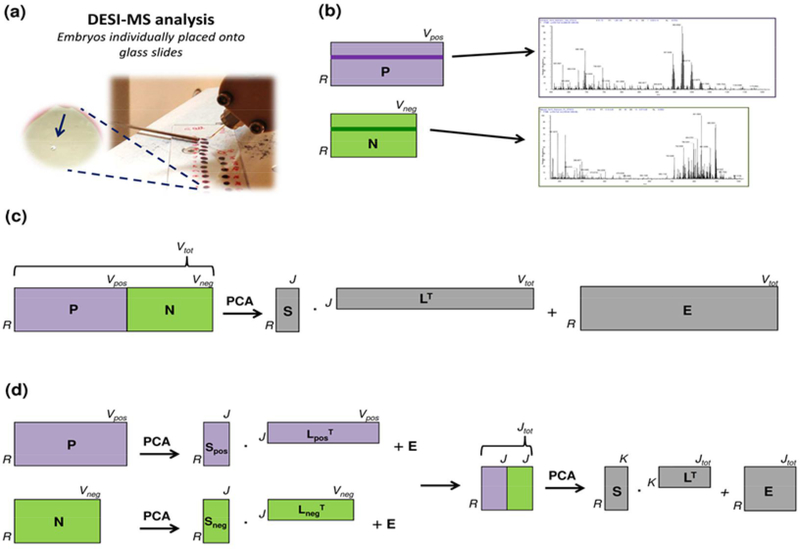
Fig. 1.
(a) DESI-MS lipid profiling of single oocytes. (b) Schematics of two matrices enclosing positive (P) and negative (N) ions for individual porcine oocytes. Positive and negative mass spectra can be visualized for the same sample by selecting the same row Ri in both of the matrices (represented as dark violet and dark green rows in P and N, respectively). (c) PCA within a low-level data fusion strategy: concatenation of two matrices with R rows (96 samples) and V variables (positive ions: 7801; negative ions: 6301) - after block-scaling to equal variance - and then computation of scores (S), loadings (L), and residuals (E). (d) PCA within a mid-level data fusion strategy: PCA was performed individually on the original datasets with R rows and V variables (positive ions: 7801; negative ions: 6301), then the same number of PCs (5) was selected as for each original dataset, and the scores of the selected PCs were merged into a new data matrix, which underwent PCA again, after column autoscaling.