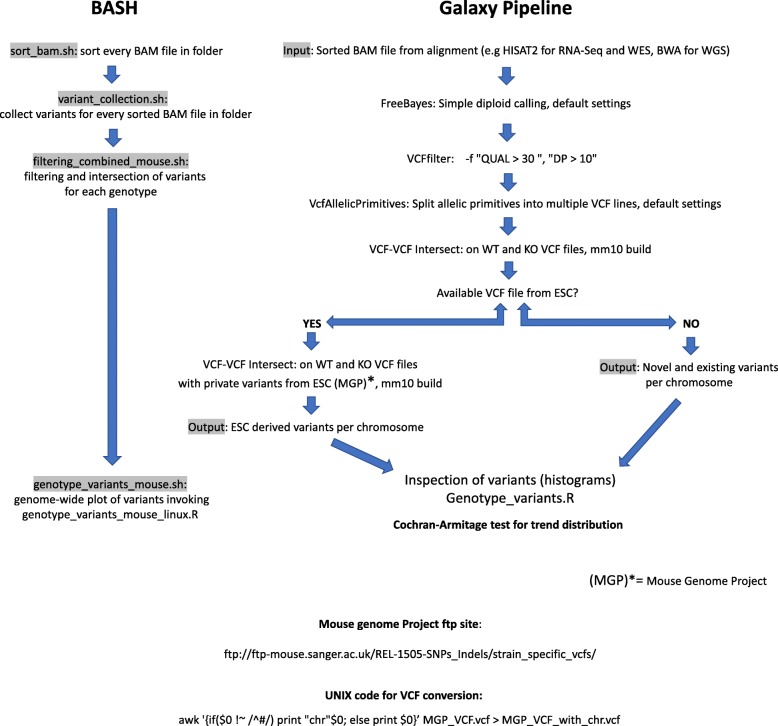
Fig. 1.
A computational pipeline for the detection of ESC-derived introgressed variants. Galaxy Platform: The pipeline starts with the input of the aligned BAM file from each genotype on the corresponding mouse genome build (e.g., HISAT2 output on the mm10 genome build for RNA-Seq data, BWA output from WES or WGS). The Freebayes variant caller program (simple variant calling) produces a VCF file from every BAM file. We filtered these VCF files using VCFlib, with the following parameters: -f “QUAL > 30”, −f “DP > 10”. Next, VCF-VCF intersect program intersects VCF files from each genotype to obtain the average variation on each genotype (mm10 build, default parameters). If the genome of the ESC used for targeting is available, and variants are correctly characterized, we can use these calls to intersect ESC introgressed variants in the VCF files from each genotype. We used VCF files available in the mouse genome project (http://www.sanger.ac.uk/science/data/mouse-genomes-project) based on the GRCm38 mouse genome release, compatible with the mm10 build (release REL-1505-SNPs_Indels). In these VCF files, the prefix “chr” in every variant call line needs to be added for compatibility with Freebayes VCF files (see UNIX code). If the genome of the ESC is not available, novel and ESC-derived variants are obtained. To confirm chromosomes with a differential distribution of variants among genotypes, we applied the Cochran-Armitage test for trend distribution. BASH: Input BAM files from RNA-Seq/WES/WGS are sorted and indexed with the sort_bam.sh script, then, variant_collection.sh script is applied for variant collection in each BAM file with Freebayes. Filtering and intersection are proceeded as described in the Galaxy platform with the filtering_combined_mouse.sh script. At this step, intersection with ESC-derived variants from the mouse project can be applied to the intersected VCF files (see Github: https://github.com/cfarkas/Genotype-variants). Finally, genome-wide plots of the intersected variants per genotype including KO-linked variants can be obtained by applying the genotype_variants_mouse.sh script