Abstract
Genome-wide scans have shown that common risk alleles for orofacial clefts (OFC) tend to be located in non-coding regulatory elements and cumulatively explain only part of the heritability of OFCs. Low-frequency variants may account for some of the “missing” heritability. Therefore, we scanned low-frequency variants located within putative craniofacial enhancers to identify novel OFC risk variants and implicate new regulatory elements in OFC pathogenesis. Analyses were performed in a multi-ethnic sample of 1995 cases of cleft lip with or without cleft palate (CL/P), 221 cases with cleft palate (CP) only, and 1576 unaffected controls. 119 putative craniofacial enhancers identified from ChIP-Seq studies in craniofacial tissues or cell lines contained multiple low-frequency (0.01% to 1%) variants, which we genotyped in participants using a custom Illumina panel. Two complementary statistical approaches, SKAT and CMC, were used to test association of the aggregated low-frequency variants across each enhancer region with CL/P and CP. We discovered a significant association between CP and a branchial arch enhancer near FOXP1 (mm60; p-value=0.0002). Additionally, we observed a suggestive association between CL/P and a forebrain enhancer near FOXE1 (hs1717; p-value=0.001). These findings suggest that low-frequency variants in craniofacial enhancer regions contribute to the complex etiology of non-syndromic OFCs.
Keywords: orofacial cleft, cleft lip, cleft palate, genetic association
Introduction
Orofacial clefts (OFCs) such as cleft lip with or without cleft palate (CL/P) and cleft palate alone (CP) are among the most common birth defects, worldwide, affecting roughly 1 in 700 births (Leslie & Marazita, 2013; Marazita, 2012). CL/P and CP are hypothesized to have distinct etiologic risk factors due to epidemiological differences and the separate embryological origins of the upper lip and secondary palate. For example, approximately 70% of CL/P and 50% of CP cases are non-syndromic, meaning they occur in the absence of other structural or cognitive abnormalities. Both non-syndromic CL/P and CP are hypothesized to be genetically complex, with contributions from multiple genetic and environmental risk factors (Leslie & Marazita, 2013).
Several genetic variants associated with OFCs have been identified through unbiased approaches, such as genome-wide association studies (GWASs) (Beaty et al., 2010; Birnbaum et al., 2009; Grant et al., 2009; Leslie, Carlson, Shaffer, Butali, et al., 2017; Leslie, Carlson, et al., 2016; Leslie, Liu, et al., 2016; Ludwig et al., 2017; Ludwig et al., 2012; Mangold et al., 2010; Sun et al., 2015; Wolf et al., 2015), which cumulatively explain about 20–30% (Ludwig et al., 2017) of the genetic variance in OFCs. These GWASs have typically interrogated common genetic variants – that is, those variants with minor allele frequencies (MAFs) greater than 1% in some human populations – due to limitations in statistical power for testing the individual effects of lower frequency variants. Altogether, results from these GWASs have suggested that many associated common variants fall in regulatory regions, which is consistent with findings from other complex diseases.
Other approaches, such as exome-wide association studies (Bureau et al., 2014; Leslie, Carlson, Shaffer, Buxo, et al., 2017) have investigated the role of low-frequency (i.e., MAF < 1%) coding variants on OFCs, implicating genes such as CDH1 (Bureau et al., 2014), N4BP2, CDSN, PRTG, and AHRR (Leslie, Carlson, Cooper, et al., 2017). Moreover, candidate gene sequencing studies have investigated the effects of rare variants (Leslie & Murray, 2013) in genes such as BMP4 (Suzuki et al., 2009), GREM1 (Al Chawa et al., 2014), and ARHGAP29 (Leslie et al., 2012). Taken together, these studies show that rare variants, both coding and regulatory, may influence risk of OFCs. Given that identifying rare variant associations is challenging, especially in non-coding regions of the genome, we hypothesize that undiscovered low-frequency variants affecting OFC risk may occur in non-coding regulatory elements, such as enhancers.
Distant-acting transcriptional enhancer regions are among the most abundant regulatory elements in the mammalian genome (Consortium, 2012; Visel, Rubin, & Pennacchio, 2009), and, from an evolutionary perspective, they may be particularly important in the emergence of human craniofacial features (Prescott et al., 2015). These enhancer regions tend to activate gene expression in specific cell or tissue types, thus providing a mechanism for the fine-tuning expression during developmental processes. Previous work in mouse has shown that craniofacial enhancers orchestrate expression of genes during craniofacial development (Attanasio et al., 2013). This leads to the question of whether genetic variation in craniofacial enhancer regions are associated with human craniofacial defects, including OFCs. Common variants in craniofacial enhancers have already been included as part of previously GWAS studies, albeit anonymously. Subsequent functional assays have demonstrated that associated SNPs at several loci disrupt the activity of their resident enhancers (Leslie et al., 2015; Lidral et al., 2015; Liu et al., 2017; Rahimov et al., 2008). However, the possibility that low-frequency variants in these regions may impact risk of OFCs has not previously been explored. In this study we scanned 119 putative craniofacial enhancer regions that harbor low-frequency genetic variants for genetic association with CL/P or CP in a multiethnic sample.
Methods
Participant recruitment
The Pittsburgh Orofacial Clefts Study (POFC) recruited a total of 11,727 participants from 11 countries across the Americas, Europe, and Asia (Leslie, Carlson, et al., 2016), which consisted of nonsyndromic orofacial cleft cases, unaffected family members of cases, and unaffected controls. For this study, we identified a subset of unrelated proband cases with orofacial clefts (1995 CL/P cases, and 221 CP cases) and 1576 unrelated controls with no history of orofacial clefts nor other craniofacial anomalies (Table 1). Cases were recruited through craniofacial clinics or surgical mission trips. Where possible, healthy controls were recruited from the same populations.
Table 1:
Number and geographic distribution of CL/P and CP cases and controls included in the analysis.
| Site | Control | CL/P case | CP case |
|---|---|---|---|
| Argentina | 30 | 111 | 20 |
| China | 27 | 157 | 23 |
| Colombia | 227 | 681 | 0 |
| Denmark | 0 | 46 | 22 |
| Guatemala | 208 | 102 | 4 |
| Hungary | 253 | 105 | 31 |
| India | 38 | 51 | 1 |
| Philippines | 96 | 159 | 16 |
| Puerto Rico | 106 | 84 | 24 |
| Spain | 0 | 34 | 4 |
| Turkey | 173 | 172 | 19 |
| United States | 418 | 293 | 57 |
| Total | 1576 | 1995 | 221 |
Editorial Policies and Ethical Considerations
All participants provided informed consent to participate in this study. All study protocols were approved by local Institutional Review Boards (IRBs) of participating sites, in addition to approvals at the University of Iowa and the University of Pittsburgh. Data analyzed in this study are publicly available at the National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGaP) study “Center for Craniofacial and Dental Genetics (CCDG): Genetic of Orofacial Clefts and Related Phenotypes” (dbGap Study Accession: phs000774.v2.p1).
Genotyping, putative enhancer selection, and imputation
Genotyping was performed using the Illumina (San Diego) HumanCore+Exome array with 15,890 single nucleotide polymorphisms (SNPs) of custom content. The custom content included SNPs in putative craniofacial enhancer sequences identified by literature search of ChIP-Seq studies in craniofacial tissues or cell lines that were available in 2013, when the custom content was selected. These included (1) enhancers from the VISTA database with in vivo activity patterns annotated for “branchial arch”, “nose”, “facial mesenchyme”, or “forebrain”; (2) putative and active enhancers identified from p300 ChIP-Seq in mouse craniofacial tissue by Attanasio et al. (Attanasio et al., 2013); and (3) craniofacial enhancers in the published literature. This field has rapidly expanded in recent years due to technological advances and epigenomic consortia such as ENCODE; therefore our selected enhancers should not be viewed as a comprehensive representation of craniofacial enhancers. Of 4,440 putative enhancer regions considered, 126 regions included multiple SNPs with MAF <5%, and, therefore, were included in our statistical analysis. Note, enhancer regions that included only one SNP were excluded from the present analysis (in which we performed aggregate tests of multiple SNPs simultaneously) based on the assumptions of our statistical approach and because tests of common variants individually have already been reported as part of previous GWAS scans (Leslie, Carlson, et al., 2016; Leslie, Liu, et al., 2016), and tests of individual low-frequency variants have insufficient statistical power. In total 804 SNPs across 126 putative craniofacial enhancer regions were analyzed (2 to 22 SNPs per enhancer; median of 5 SNPs). Sporadic missing genotypes were imputed using IMPUTE2 (Howie, Donnelly, & Marchini, 2009) software with the 1000 Genomes Project (phase 3 release) as the reference panel. Imputation accuracy was assessed by masked variant analysis, which showed high quality imputation with mean concordance of 0.995. Most-likely genotypes were used in statistical analysis if the genotype probability was greater than 0.9, an arbitrary threshold set to ensure high confidence in imputed data.
Statistical analysis
Genetic association of aggregated low-frequency variants across putative craniofacial enhancer regions was tested using two approaches: the combined multivariate and collapsing (CMC) method and the sequence kernel association test (SKAT). In brief, CMC is a burden test that collapses variants into a single score while assuming consistent direction of effect of minor alleles of all SNPs in an enhancer region (Li & Leal, 2008). In contrast, SKAT allows minor alleles of SNPs to differ in the direction of effect (M. C. Wu et al., 2011). These two approaches are complementary in that each has higher power in some circumstances, depending on whether or not minor alleles of SNPs with the enhancer region truly have effects in the same direction.
Tests of enhancer regions using CMC and SKAT were performed separately for CL/P and CP phenotypes as implemented in RVTESTS (Zhan, Hu, Li, Abecasis, & Liu, 2016) software. Two MAF ranges for low-frequency variants were considered. The principal analyses included putative enhancer regions with multiple low-frequency variants in the 0.01% to 1% range. For secondary analyses, this range was expanded include variants up to MAF of 5%. Because case-control analyses of CL/P and CP comprised different sets of participants, minor alleles were not observed for all variants in both analysis groups, and therefore not all enhancer regions were tested for each phenotype. Specifically, for the analyses of low-frequency variants with MAF <1%, the CL/P scan tested 669 SNPs across 118 regions, whereas the CP scan tested 586 variants across 108 regions. For the analyses of variants with MAF <5%, the CL/P scan tested 804 SNPs across 126 regions, and the CP scan tested 719 SNPs across 115 regions.
To adjust for population structure, we included the first 18 principal components (PCs) of ancestry in our models as covariates. PCs of ancestry were derived from genome-wide data as previously described (Leslie, Carlson, et al., 2016). Empirical significance for SKAT models was determined by 10,000 permutations. Associations with p-values less than the Bonferroni-corrected (for the number of regions tested) threshold were considered statistically significant, and p-values less than 0.004 (i.e., roughly one order of magnitude more liberal than Bonferroni adjustment) were considered suggestive associations. For enhancer regions showing evidence of association, we scrutinized the quality of genotype calling by inspecting clustering in allele intensity plots. In addition, for enhancer regions showing evidence of association via CMC or SKAT we performed SNP-wise tests for all variants in the enhancer region using logistic regression. SNPs showing individual effects were interrogated using HaploReg for chromatin state, protein binding, and eQTL annotations. Localization of enhancer activity was depicted using representative images from The VISTA Enhancer Browser (Visel, Minovitsky, Dubchak, & Pennacchio, 2007).
Results
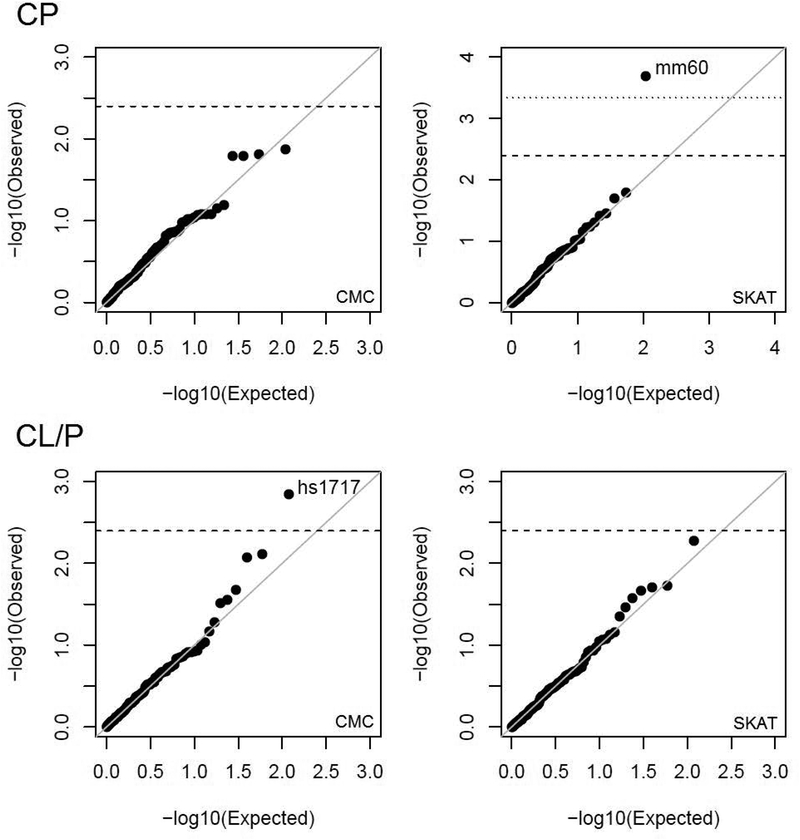
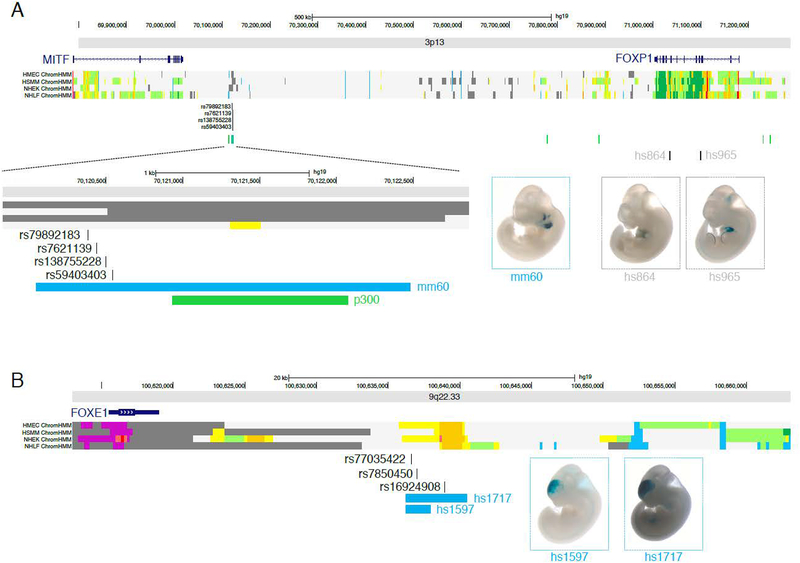
Low-frequency variants (MAF 0.01% to 1%) were tested in aggregate using two methods (CMC and SKAT) for putative craniofacial enhancer regions. Results are summarized in quantile-quantile plots shown in Figure 1. One association (SKAT p-value = 2.0 × 10−4; CMC p-value = 0.08) was observed between CP and a branchial arch enhancer, mm60, located on chromosome 3p13 (position 70,120,048–70,122,477 [hg19], Figure 2A) between MITF (103kb from enhancer) and FOXP1 (883kb from enhancer). No significantly associated enhancer regions were observed for CL/P, however, hs1717, an enhancer region on chromosome 9q22.33 (position 100636317–100639947, Figure 2B) near FOXE1 (20kb from enhancer), showed suggestive evidence (SKAT p-value 0.005; CMC p-value = 0.001). The activity patterns in E11.5 mouse are localized to the first and second branchial arches for mm60, and to the forebrain for hs1717 (Figure 2C–D).
Figure 1: Results of genetic association tests:
Quantile-quantile plots depict the observed log10-transformed p-values (y-axis) vs. the expected distribution of p-values (x-axis) under the null hypothesis of no association. Each point represents the evidence of genetic association for a specific craniofacial enhancer. The top panels show results for CP and the bottom panels show results for CL/P. Left panels show results of CMC scans, and right panels show results of SKAT scans. Horizontal dashed lines represent the threshold for suggestive association (p-value < 0.004). The horizontal dotted line represents the Bonferroni threshold for statistical significance.
Figure 2: Genomic position and activity localization of associated enhancers:
(A) Position of the chromosome 3p13 craniofacial enhancer, mm60, and possible cis-regulatory target genes MITF and FOXP1. Chromatin states in normal human cell lines (dermal endothelium [HMEC], skeletal muscle [HSMM], epidermal keratinocytes [NHEK], and lung fibroblasts [NHLF]) are colored-coded with yellow and orange segments representing putative enhancer regions. The associated craniofacial enhancer, mm60, is active in the branchial arches at E11.5, and does not show enhancer chromatin states in cell lines. Other enhancers near FOXP1 (hs864, hs965) not associated with OFCs are active in the developing heart and limb, consistent with the expression pattern of FOXP1. (B) Position of the chromosome 9q22.33 craniofacial enhancer, hs1717, and possible cis-regulatory target gene FOXE1. The hs1717 enhancer can be subdivided into a smaller enhancer (hs1597), each showing activity in the forebrain. Enhancer activity of hs1717 is consistent with chromatin states in human cell lines.
Table 2 shows the SNP-wise association test results for low-frequency variants in the significant mm60 and suggestive hs1717 enhancer regions. Effects of individual SNPs suggest multiple variants may be contributing to the associations, with one SNP per locus showing statistically significant individual effects. Inspection of genotype clustering plots did not indicate any genotyping problems for these loci (results not shown). For the low-frequency SNPs in the mm60 enhancer, rs138755228 was statistically the most significantly associated with CP (p-value=0.008); this SNP is located in a region annotated with chromatin state and histone modifications consistent with enhancer activity in multiple cell types, but is not a recognized eQTL. For the low-frequency SNPs in the hs1717 enhancer, rs16924908 was the most significant (p-value=0.02), and also shows chromatin state and histone marks consistent with its enhancer activity across multiple cell types, and is also not a recognized eQTL.
Table 2:
Results of single SNP tests of association with CP at enhancer mm60 (chromosome 3p13) and with CL/P at enhancer hs1717 (chromosome 9q22.33).
| Enhancer | Chromosome | SNP | BP (hg19) | MAF controls | MAF cases | Odds ratio | P-value |
|---|---|---|---|---|---|---|---|
| mm60 | 3p13 | rs59403403 | 70120542 | 0.003383 | 0 | NA | 0.2207 |
| rs7621139 | 70120438 | 0.002153 | 0 | NA | 0.3289 | ||
| rs79892183 | 70120383 | 0.000923 | 0 | NA | 0.5229 | ||
| rs138755228 | 70120496 | 0 | 0.002262 | NA | 0.006671 | ||
| hs1717 | 9q22.33 | rs77035422 | 100636618 | 0.00123 | 0.002005 | 1.631 | 0.4199 |
| rs7850450 | 100636981 | 0.003383 | 0.001754 | 0.5178 | 0.1663 | ||
| rs16924908 | 100638923 | 0.01046 | 0.005514 | 0.5248 | 0.01695 |
To expand the scope of the low-frequency variant scans, we re-analyzed putative enhancer regions while also considering more common variants of up to 5% MAF. Results for CMC and SKAT tests of the broader group low-frequency variants (MAF 0.01% to 5%) did not identify any additional significant associations, but did point to one new suggestive enhancer region (Supplemental Figure S1). However, inspection of allele intensity plots for SNPs in this suggestive enhancer region determined that the association may have been driven by a SNP with poor genotype clustering due to a null allele (Supplemental Figure S2). We interpret this suggestive association with great skepticism, and therefore discuss it no further.
Discussion
In this study we tested low-frequency variants in over 100 putative craniofacial enhancers across the genome for association with CL/P and CP. We identified an enhancer, mm60, near MITF and FOXP1 that was significantly associated with CP. Neither of these genes have been previously linked to OFCs, although both genes may be involved in craniofacial phenotypes.
MITF is a transcription factor involved in multiple cellular processes; notably, mutations in MITF cause Waardenburg syndrome, characterized by pigment abnormalities, hearing loss, telecanthus, and other symptoms including cleft lip that vary across patients. However, most, but not all (Pierpont, St Jacques, Seaver, & Erickson, 1995), cases of Waardenburg syndrome presenting with clefts are due to mutations in another gene, PAX3, which has been further supported by a cleft palate murine model (M. Wu et al., 2008). To our knowledge, clefts have not been documented in Waardenburg syndrome cases caused by mutations in MITF.
FOXP1 is a transcription factor important for regulating gene expression during development. Mutations in FOXP1 are associated with developmental delay, intellectual disability, speech defects, and mild craniofacial abnormalities (Horn, 2012; Le Fevre et al., 2013; Myers et al., 2017), including highly arched palate and hypertrophy of the alveolar ridges (Urreizti et al., 2018). In mouse, expression patterns of Foxp1 in the first pharyngeal arch during development suggest that it may regulate jaw development, although expression was absent from the palatal shelf (Cesario, Almaidhan, & Jeong, 2016). In addition, knockout mutants for Lhx6 and Lhx8, which bind to and repress transcription of FOX genes, show up-regulation of Foxp1 (as well as Foxp2, Foxc1, Foxd1, and Foxd2) and have major craniofacial defects including cleft palate (Cesario et al., 2015). Taken together, these studies suggest that FOXP1 may be involved in regulating craniofacial development. Furthermore, if the chromosome 3p13 enhancer near FOXP1 regulates its expression, our results would point to a role of FOXP1 in risk of CP, although more work is necessary to replicate this association. We advocate that FOXP1 should be considered a high-priority target for future genetics studies of OFCs, and CP in particular, the cleft subtype for which fewer specific genes have been discovered.
In addition to the significant association observed for CP, we also identified an enhancer, hs1717, near FOXE1 that was suggestively associated with CL/P, but this relationship was not statistically significant after consideration of multiple testing. We observed similar effects in stratified analyses of both cleft lip alone and cleft lip and palate (results not shown). FOXE1 is a transcription factor expressed during palate formation and has been implicated in OFCs via multiple genetic approaches, including linkage analyses (Marazita et al., 2009; Moreno et al., 2009), candidate gene sequencing studies (Moreno et al., 2009), and GWAS (Leslie, Carlson, Shaffer, Butali, et al., 2017). The fact that we observe statistical evidence for an enhancer near this well-established OFC gene lends some credence to our study design, serving as an informal positive control.
Two rare variant tests were used, CMC and SKAT, because these approaches each have greater statistical power in some circumstances. CMC, which collapses the burden of low-frequency variants into a single score, operates under the assumption that all low-frequency alleles in an enhancer region are deleterious, whereas SKAT can detect association with sets of variants when the low-frequency alleles differ in the directions of their effects. As we do not know the directions of effects ahead of time, CMC and SKAT approaches complement each other in our efforts to scan craniofacial enhancer regions across the genome for evidence of association with cleft phenotypes. This approach is commonly employed in rare variant analyses, and has proved useful in our study for detecting significant and suggestive associations.
This study was carried out in a multi-ethnic sample of participants, which included individuals of European, Asian, and South American ancestry. While spurious associations due to population structure were avoided by thoroughly adjusting for dimensions of genetic ancestry, it is possible that the rare-variant associations observed are driven primarily by one or more ethnic populations in the total sample, as allele frequencies, especially for rare variants, may differ across populations. Unfortunately, sample sizes were too small to perform rare variants tests separately by ancestry group. Larger and more homogeneous samples and sequencing approaches may be necessary to find any associations that went undetected in this analysis.
Additional studies will be needed to confirm the impact of the mm60 and hs1717 enhancer regions on OFCs, which poses a challenge in that the large sample sizes necessary for detecting low-frequency variants of modest effects are costly to produce. Systems genetics approaches, which combine genotype information with other ‘omics data, may be helpful in overcoming this challenge. Whereas the present study utilized a two-stage approach, whereby prior ChIP-Seq experiments were used to select high-priority, putative craniofacial enhancer regions to be investigated in independent genetic analyses, fully integrative approaches that combine information on genetic variants with data from expression, methylation, and/or protein-binding experiments may assist in unraveling the molecular basis of OFC risk. Based on our findings, investigations of craniofacial enhancer regions may be fruitful in identifying OFC risk variants, especially as detailed catalogs of regulatory elements and whole genome sequencing becomes available on a large scale. Given that enhancer detection and annotation is currently a rapidly developing area of science, the set of 4,440 putative craniofacial enhancer regions considered in this study may be viewed as a preliminary list; future efforts in this vein may benefit from an expanded and refined catalog of craniofacial enhancers as this field matures. Though results from this study are unlikely to have any direct clinical benefit at this time, ultimately, knowledge of genetic architecture of OFCs may be beneficial for risk profiling and recurrence prediction.
Supplementary Material
Acknowledgements
We gratefully acknowledge the participation of the families, field staff, and collaborators around the world who made this project possible. In particular we highlight the contributions of Dr. Andrew Czeizel (deceased), Dr. Juan C. Mereb (deceased), and Eduardo Castilla (deceased). This work was supported by grants from the National Institutes of Health (NIH) including: R00-DE025060 [EJL], X01-HG007485 [MLM, EF], R01-DE016148 [MLM, SMW], U01-DE024425 [MLM], R37-DE008559 [JCM, MLM], R01-DE009886 [MLM], R21-DE016930 [MLM], R01-DE014667 [LM], R01-DE012472 [MLM], R01-DE011931 [JTH], R01-DE011948 [KC], U01-DD000295 [GLW], R25-MD007607 [CJB], K99-DE024571 [CJB], S21-MD001830 [CJB], U54-MD007587 [CJB]. Genotyping and data cleaning were provided via an NIH contract to the Johns Hopkins Center for Inherited Disease Research: HHSN268201200008I. Additional support provided by: the National Science Foundation Grant DBI-1263020, which was co-funded by the Department of Defense; the Robert Wood Johnson Foundation, AMFDP Grant 72429 [AB]; an intramural grant from the Research Institute of the Children’s Hospital of Colorado [FWD]; operating costs support in the Philippines was provided by the Institute of Human Genetics, National Institutes of Health, University of the Philippines, Manila [CP]; grants through FAPERJ, Brazil [IMO]: grant numbers: E-26/20300/2017; grants through CNPq, Brazil [IMO]: grant numbers: 310772/2017–6, 400427/2013–3.
Footnotes
Conflict of Interest
All authors have no conflicts of interest to disclose.
References
- Al Chawa T, Ludwig KU, Fier H, Potzsch B, Reich RH, Schmidt G, … Mangold E (2014). Nonsyndromic cleft lip with or without cleft palate: Increased burden of rare variants within Gremlin-1, a component of the bone morphogenetic protein 4 pathway. Birth Defects Res A Clin Mol Teratol, 100(6), 493–498. doi: 10.1002/bdra.23244 [DOI] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, … Visel A (2013). Fine tuning of craniofacial morphology by distant-acting enhancers. Science, 342(6157), 1241006. doi: 10.1126/science.1241006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, … Scott AF (2010). A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet, 42(6), 525–529. doi: 10.1038/ng.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, … Mangold E (2009). Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet, 41(4), 473–477. doi: 10.1038/ng.333 [DOI] [PubMed] [Google Scholar]
- Bureau A, Parker MM, Ruczinski I, Taub MA, Marazita ML, Murray JC, … Beaty TH (2014). Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics, 197(3), 1039–1044. doi: 10.1534/genetics.114.165225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario JM, Almaidhan AA, & Jeong J (2016). Expression of forkhead box transcription factor genes Foxp1 and Foxp2 during jaw development. Gene Expr Patterns, 20(2), 111–119. doi: 10.1016/j.gep.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario JM, Landin Malt A, Deacon LJ, Sandberg M, Vogt D, Tang Z, … Jeong J (2015). Lhx6 and Lhx8 promote palate development through negative regulation of a cell cycle inhibitor gene, p57Kip2. Hum Mol Genet, 24(17), 5024–5039. doi: 10.1093/hmg/ddv223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489(7414), 57–74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, … Hakonarson H (2009). A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr, 155(6), 909–913. doi: 10.1016/j.jpeds.2009.06.020 [DOI] [PubMed] [Google Scholar]
- Horn D (2012). Mild to Moderate Intellectual Disability and Significant Speech and Language Deficits in Patients with FOXP1 Deletions and Mutations. Mol Syndromol, 2(3–5), 213–216. doi:000330916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, & Marchini J (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet, 5(6), e1000529. doi: 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fevre AK, Taylor S, Malek NH, Horn D, Carr CW, Abdul-Rahman OA, … Hunter MF (2013). FOXP1 mutations cause intellectual disability and a recognizable phenotype. Am J Med Genet A, 161A(12), 3166–3175. doi: 10.1002/ajmg.a.36174 [DOI] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Cooper ME, Christensen K, Weinberg SM, & Marazita ML (2017). Exploring Subclinical Phenotypic Features in Twin Pairs Discordant for Cleft Lip and Palate. Cleft Palate Craniofac J, 54(1), 90–93. doi: 10.1597/15-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Butali A, Buxo CJ, Castilla EE, … Marazita ML (2017). Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum Genet, 136(3), 275–286. doi: 10.1007/s00439-016-1754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Buxo CJ, Castilla EE, Christensen K, … Marazita ML (2017). Association studies of low-frequency coding variants in nonsyndromic cleft lip with or without cleft palate. Am J Med Genet A, 173(6), 1531–1538. doi: 10.1002/ajmg.a.38210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, … Marazita ML (2016). A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet, 25(13), 2862–2872. doi: 10.1093/hmg/ddw104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Liu H, Carlson JC, Shaffer JR, Feingold E, Wehby G, … Marazita ML (2016). A Genome-wide Association Study of Nonsyndromic Cleft Palate Identifies an Etiologic Missense Variant in GRHL3. Am J Hum Genet, 98(4), 744–754. doi: 10.1016/j.ajhg.2016.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Mansilla MA, Biggs LC, Schuette K, Bullard S, Cooper M, … Murray JC (2012). Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res A Clin Mol Teratol, 94(11), 934–942. doi: 10.1002/bdra.23076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, & Marazita ML (2013). Genetics of cleft lip and cleft palate. Am J Med Genet C Semin Med Genet, 163C(4), 246–258. doi: 10.1002/ajmg.c.31381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, & Murray JC (2013). Evaluating rare coding variants as contributing causes to non-syndromic cleft lip and palate. Clin Genet, 84(5), 496–500. doi: 10.1111/cge.12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Taub MA, Liu H, Steinberg KM, Koboldt DC, Zhang Q, … Murray JC (2015). Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am J Hum Genet, 96(3), 397–411. doi: 10.1016/j.ajhg.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, & Leal SM (2008). Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet, 83(3), 311–321. doi: 10.1016/j.ajhg.2008.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Liu H, Bullard SA, Bonde G, Machida J, Visel A, … Cornell RA (2015). A single nucleotide polymorphism associated with isolated cleft lip and palate, thyroid cancer and hypothyroidism alters the activity of an oral epithelium and thyroid enhancer near FOXE1. Hum Mol Genet, 24(14), 3895–3907. doi: 10.1093/hmg/ddv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Leslie EJ, Carlson JC, Beaty TH, Marazita ML, Lidral AC, & Cornell RA (2017). Identification of common non-coding variants at 1p22 that are functional for non-syndromic orofacial clefting. Nat Commun, 8, 14759. doi: 10.1038/ncomms14759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Bohmer AC, Bowes J, Nikolic M, Ishorst N, Wyatt N, … Mangold E (2017). Imputation of orofacial clefting data identifies novel risk loci and sheds light on the genetic background of cleft lip +/− cleft palate and cleft palate only. Hum Mol Genet, 26(4), 829–842. doi: 10.1093/hmg/ddx012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, … Nothen MM (2012). Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet, 44(9), 968–971. doi: 10.1038/ng.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, … Nothen MM (2010). Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet, 42(1), 24–26. doi: 10.1038/ng.506 [DOI] [PubMed] [Google Scholar]
- Marazita ML (2012). The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet, 13, 263–283. doi: 10.1146/annurev-genom-090711-163729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Lidral AC, Murray JC, Field LL, Maher BS, Goldstein McHenry T, … Arcos-Burgos M (2009). Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Hum Hered, 68(3), 151–170. doi: 10.1159/000224636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno LM, Mansilla MA, Bullard SA, Cooper ME, Busch TD, Machida J, … Lidral AC (2009). FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum Mol Genet, 18(24), 4879–4896. doi: 10.1093/hmg/ddp444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A, du Souich C, Yang CL, Borovik L, Mwenifumbo J, Rupps R, … Boerkoel CF (2017). FOXP1 haploinsufficiency: Phenotypes beyond behavior and intellectual disability? Am J Med Genet A, 173(12), 3172–3181. doi: 10.1002/ajmg.a.38462 [DOI] [PubMed] [Google Scholar]
- Pierpont JW, St Jacques D, Seaver LH, & Erickson RP (1995). A family with unusual Waardenburg syndrome type I (WSI), cleft lip (palate), and Hirschsprung disease is not linked to PAX 3. Clin Genet, 47(3), 139–143. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7634536 [DOI] [PubMed] [Google Scholar]
- Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, … Wysocka J (2015). Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell, 163(1), 68–83. doi: 10.1016/j.cell.2015.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, … Murray JC (2008). Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet, 40(11), 1341–1347. doi: 10.1038/ng.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, … Yang Y (2015). Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun, 6, 6414. doi: 10.1038/ncomms7414 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A, … Murray JC (2009). Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am J Hum Genet, 84(3), 406–411. doi: 10.1016/j.ajhg.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urreizti R, Damanti S, Esteve C, Franco-Valls H, Castilla-Vallmanya L, Tonda R, … Balcells S (2018). A De Novo FOXP1 Truncating Mutation in a Patient Originally Diagnosed as C Syndrome. Sci Rep, 8(1), 694. doi: 10.1038/s41598-017-19109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, & Pennacchio LA (2007). VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res, 35(Database issue), D88–92. doi: 10.1093/nar/gkl822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, & Pennacchio LA (2009). Genomic views of distant-acting enhancers. Nature, 461(7261), 199–205. doi: 10.1038/nature08451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf ZT, Brand HA, Shaffer JR, Leslie EJ, Arzi B, Willet CE, … Bannasch DL (2015). Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet, 11(3), e1005059. doi: 10.1371/journal.pgen.1005059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Li J, Engleka KA, Zhou B, Lu MM, Plotkin JB, & Epstein JA (2008). Persistent expression of Pax3 in the neural crest causes cleft palate and defective osteogenesis in mice. J Clin Invest, 118(6), 2076–2087. doi: 10.1172/JCI33715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, & Lin X (2011). Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet, 89(1), 82–93. doi: 10.1016/j.ajhg.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Hu Y, Li B, Abecasis GR, & Liu DJ (2016). RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics, 32(9), 1423–1426. doi: 10.1093/bioinformatics/btw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.