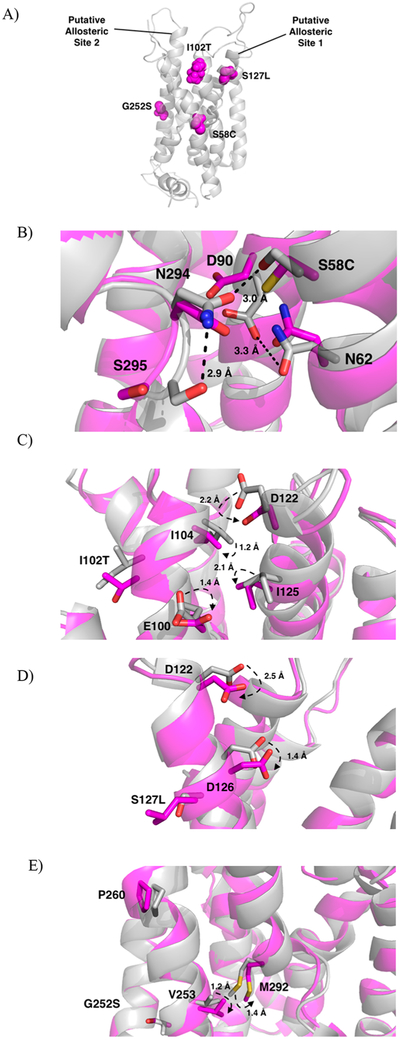
Figure 4.
(A) Location of the mutated residues (magenta, spheres) within the protein (gray, cartoon) and labeled putative allosteric sites. (B-E) Overlap of wild-type (gray) and mutated proteins (magenta). (B) Mutation of Ser58 to Cys58 causes disruption of the H-bond network stabilizing TM6. Hydrogen bonds with distances in the wild-type protein shown as dashed lines (black). Distances increase between N294 and S295 from 2.9 to 4.2 Å, between N294 and S58 from 3.0 to 3.2 Å, and between D90 and N62 from 3.3 to 5.3 Å. Distances between residues upon mutation not shown for clarity. (C) Mutation of Ile102 to Thr102 disrupts the TM2-TM3 interface imperative for receptor activation. (D) Mutation of S127 to Leu127 causes a shift in Asp122 and Asp126, critical residues for ligand recognition. (E) Mutation of Gly252 to Ser252 interrupts helix-helix interactions necessary for stabilization of TM6.