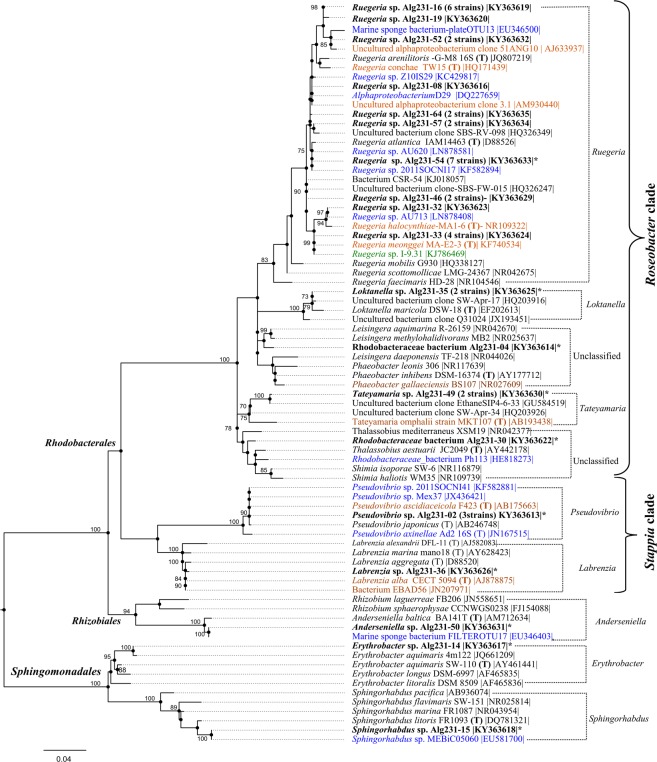
Figure 1.
16S rRNA gene Maximum Likelihood tree of Alphaproteobacteria species. Kimura 2-parameter evolutionary distances between sequences were calculated using MEGA792. Alphaproteobacteria strains isolated from S. officinalis are shown in bold, with each entry representing a unique OTU at 100% nucleotide homology cut-off. The number of isolates obtained from S. officinalis that belong to the same OTU are given in brackets. Closest NCBI BlastN hits and type strains (T) to each isolate are shown on the tree. Blue marks sponge-associated, orange marks invertebrate-associated and green marks marine algae-associated closest NCBI BlastN hits and type strains. Strains that had their genome sequenced are marked with an asterisk. Bootstrap values (500 repetitions) above 70% (0.7) are shown on the tree nodes. The tree contains 80 entries, and 682 nucleotide positions are included in the dataset.