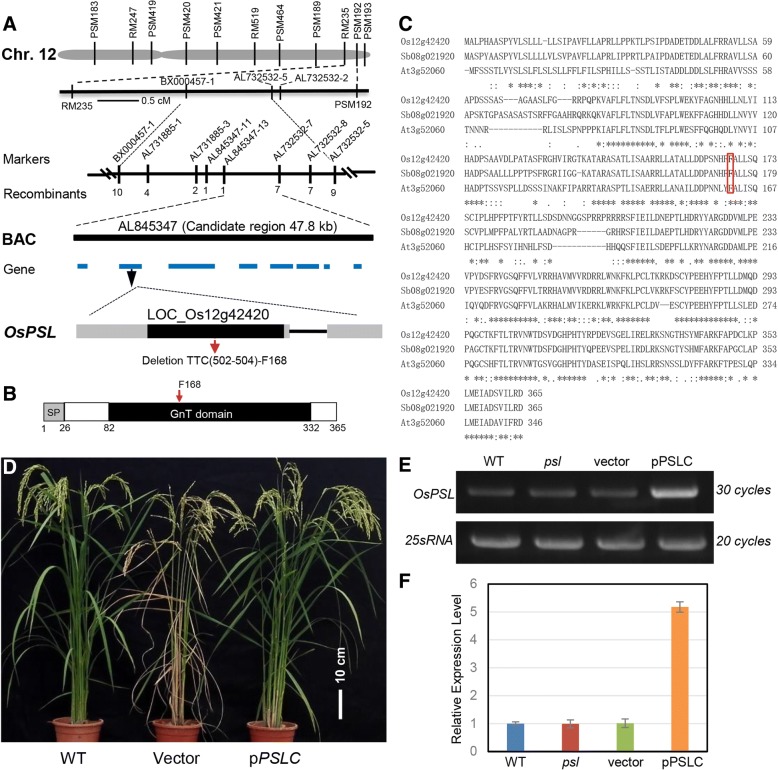
Fig. 4.
Map-based cloning of the OsPSL locus. a The PSL locus was mapped to the long arm of rice chromosome 12. Black boxes indicate the coding sequence, grey boxes indicate the 5′ and 3′untranslated regions, and lines between boxes indicate introns. Deletion site identified in the psl are indicated by a red arrowhead. b Schematic representation of the OsPSL protein domains. Positions of amino acid residues delimiting each of the indicated domains are shown. c Amino acid sequence alignment of the rice OsPSL (Os12g42420) with its homologs from Arabidopsis. thaliana (At3g52060) and Sorghum. bicolor (sb08g021920). Symbol designations: “*” identical residues, “:” conserved substitutions, “.” semi-conserved substitutions. The conserved phenylalanine residue likely involved in the enzymatic activity is indicated by a red box, and gaps introduced for alignment are indicated by dashes. d Complementation analysis of the psl mutant. Plant phenotype of WT and psl mutant transformed with OsPSL (pPSLC) or with empty vector (Vector) grown in a paddy field. Expression analysis of OsPSL gene in WT, psl mutant and the transgenic plants with OsPSL (pPSLC) or with empty vector (Vector) by semi-quantitative (e) and quantitative (f) RT-PCR