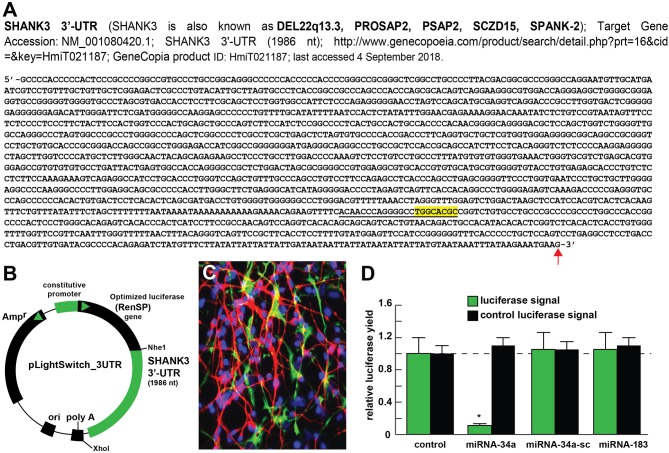
Figure 2.
Luciferase reporter vector-based studies of miRNA-34a and SHANK3 expression - Functional validation of a miRNA-34a-SHANK3–3′UTR interaction. (A) partial ribonucleotide sequence of the 1986 nt SHANK3-mRNA-3′-UTR is shown in the 5′-3′ direction; the 22 nucleotide (nt) miRNA-34a-SHANK3-3′UTR complementarity-interaction region is indicated by a black underline and the 8 nt SHANK3-mRNA-3′-UTR seed sequence is overlaid in yellow; a single vertical red arrow indicates the 5′ end of a poly A+ tail in the SHANK3 mRNA; the SHANK3 mRNA sequence derived from NM_018965; (B) SHANK3-mRNA-3′UTR expression vector luciferase reporter assay (pLightSwitch-3′UTR; Cat#S801178; Switchgear Genomics, Palo Alto CA); in this vector, the entire 1986 nucleotide SHANK3 3′UTR was ligated into the unique Nhe1-Xho1site; not drawn to scale; (C) HNG cells, 2 weeks in primary culture; neurons (red stain; λmax = 690 nm), DAPI (blue nuclear stain; λmax = 470 nm) and glial fibrillary associated protein (GFAP; glial-specific green stain; λmax = 520 nm); the HNG cell culture is about 60% confluent and at 2 weeks of culture contains 70% neurons and 30% astroglia; human neurons do not culture well in the absence of glia; neurons also show both extensive arborization and display electrical activity (unpublished; Lonza); 40X magnification; HNG cells were transfected with the SHANK3-mRNA-3′UTR expression vector luciferase reporter were treated exogenously with a stabilized miRNA-34a, a scrambled control miRNA-34a (miRNA-34a-sc) or control miRNA-183; see references and text for further details; (D) compared to control, HNG cells transfected with a scrambled (sc) control pLightSwitch-3'UTR vector, the SHANK3-mRNA-3′UTR vector exhibited decreased luciferase signal to a mean of 0.16-fold of controls in the presence of miRNA-34a; this same vector exhibited no change in the presence of the control miRNA-34a-sc or miRNA-183; for each experiment (using different batches of HNG cells) a control luciferase signal was generated and included separate controls with each analysis; in addition a control vector β-actin-3′UTR showed no significant effects on the relative luciferase signal yield after treatment with either miRNA-183 or miRNA-34a (data not shown); a dashed horizontal line set to 1.0 is included for ease of comparison; N = 6; *p < 0.001 (ANOVA). The results suggest a physiologically relevant miRNA-34a-SHANK3-mRNA-3′UTR interaction and a miRNA-34a-mediated down-regulation of SHANK3 expression in HNG cells. This pathogenic interaction may be related to the down-regulation of other immune, inflammatory, and synaptic system genes by up-regulated miRNAs in the CNS resulting in an impairment in trans-synaptic signaling and synaptic cytoarchitecture.