Figure 5.
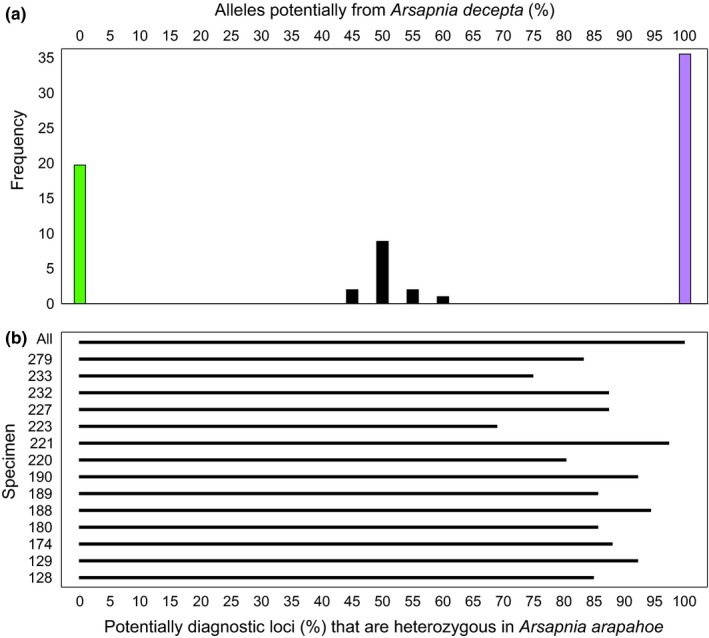
Patterns of genome‐wide SNPs (n = 94) in Arsapnia arapahoe (n = 14; black bars) that were fixed for alternate (and potentially diagnostic) alleles in Capnia gracilaria (n = 20; green bars) and A. decepta (n = 36; purple bars) in samples of these species from Colorado. (a) The percentage of alleles potentially diagnostic for A. decepta. That value subtracted from 100 equals the percentage of loci potentially derived from Capnia gracilaria. The central peak represents A. arapahoe with equal contributions of alleles from both potential parental species. (b) The percentage of loci at which A. arapahoe specimens are heterozygous. For 60 presumably autosomal loci, all A. arapahoe specimens (all) were heterozygous at all loci, indicating that alleles at these loci are diagnostic. For an additional 34 nondiagnostic autosomal SNPs, percentages of loci that were heterozygous are given for each specimen. Not shown are results for 51 sex‐linked SNPs for which all male A. arapahoe (specimens 128–233) are fixed for the alleles from A. decepta. The single female A. arapahoe (specimen 279) was heterozygous for each of these loci
