Fig. 1.
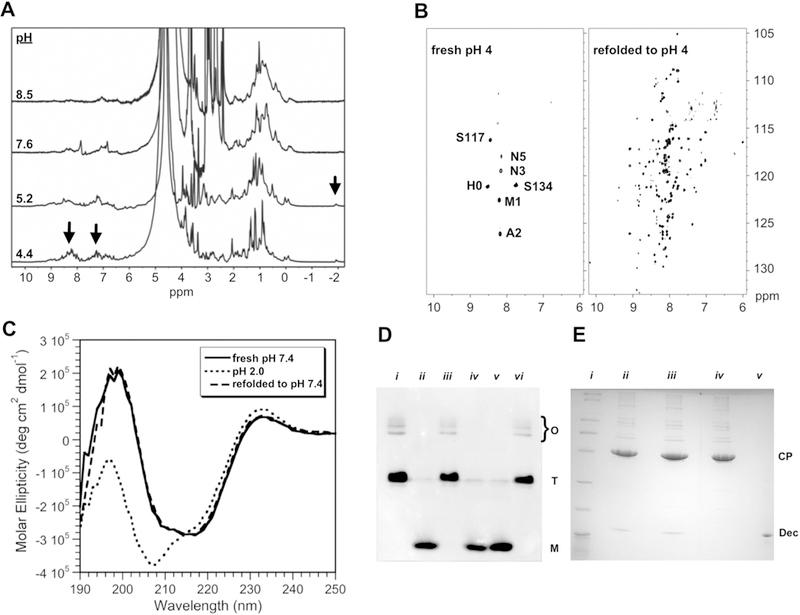
Optimization of NMR conditions. a 1D 1H NMR spectra showing narrowing of signals (arrows) at low pH. b Comparison of 600 MHz 1H–15N TROSY spectra of freshly prepared Dec at pH 4, with Dec that was unfolded at pH 2 for 20 min before refolding to pH 4. c CD experiments to verify that the refolding protocol does not change the secondary structure of Dec. These spectra are compared with Dec at pH 2, which has a CD spectrum typical of an unfolded protein. CD spectra were performed as previously described (Parent et al. 2012), using 0.45 mg/ml Dec in 10 mM NaPO4 at 33 °C. d Native (15% polyacrylamide) gel electrophoresis showing that unfolding of Dec to pH 2 followed by refolding to higher pH, converts it from a trimer to a monomer. Monomer (M), trimer (T), and bands from higher-order oligomers (O) are indicated. Lanes: (i) Dec at pH 7.6; (ii) Dec unfolded at pH 2, followed by refolding to pH 7.6.; (iii) Dec at pH 4; (iv) Dec at pH 4 after unfolding to pH 2 and refolding; (v) Dec in its acid-unfolded state at pH 2; (vi) Lowering the pH from 7.6 to 4 does not convert Dec from a trimer to a monomer (the protein needs to be unfolded to pH 2 to obtain the monomer). e Experiment to show Dec remains competent to bind P22 capsids after unfolding at pH 2, followed by refolding to pH 7.4. Dec samples were mixed with P22 phage and the complexes were separated from unbound Dec by a CsCl gradient, followed by TCA precipitation and analysis by 15% SDS-PAGE. The positions of the Dec and CP (phage coat protein) bands are indicated. Lanes: (i) MW markers (BioRad broad-range SDS-PAGE standards #161–0737). (ii) Native Dec bound to P22 phages at pH 7.4. (iii) Dec retains its ability to bind P22 after refolding from pH 2 to pH 7.4. (iv) P22 phages alone. (v) Dec protein marker
