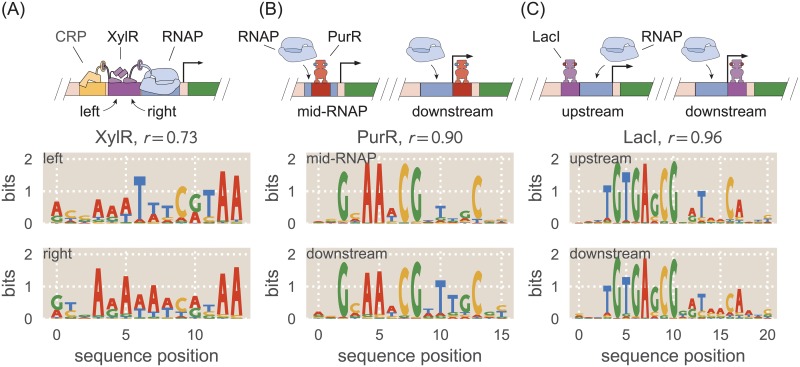
Fig 6. Regulatory context can alter sequence preference.
Sequence logos were obtained for the same transcription factors in different regulatory contexts and compared against one another. The Pearson’s correlation coefficient r between energy matrices is noted for each pair of binding sites. A: Sequence logos are shown for the two adjacent binding sites for the activator XylR in the xylE promoter, shown schematically at top. The sequence logos for the two binding sites indicate that they have significantly different sequence preferences. B: Sequence logos are shown for the PurR binding site in the purT promoter and a PurR binding site for a synthetic simple repression promoter in which the binding site is positioned differently, shown schematically at top. The sequence logos for the two binding sites indicate nearly identical sequence preferences. C: Sequence logos are shown for a LacI binding site upstream of the RNAP binding site and a LacI binding site downstream of the RNAP. Although regulatory mechanisms differ between these two binding sites, their sequence logos are nearly identical.