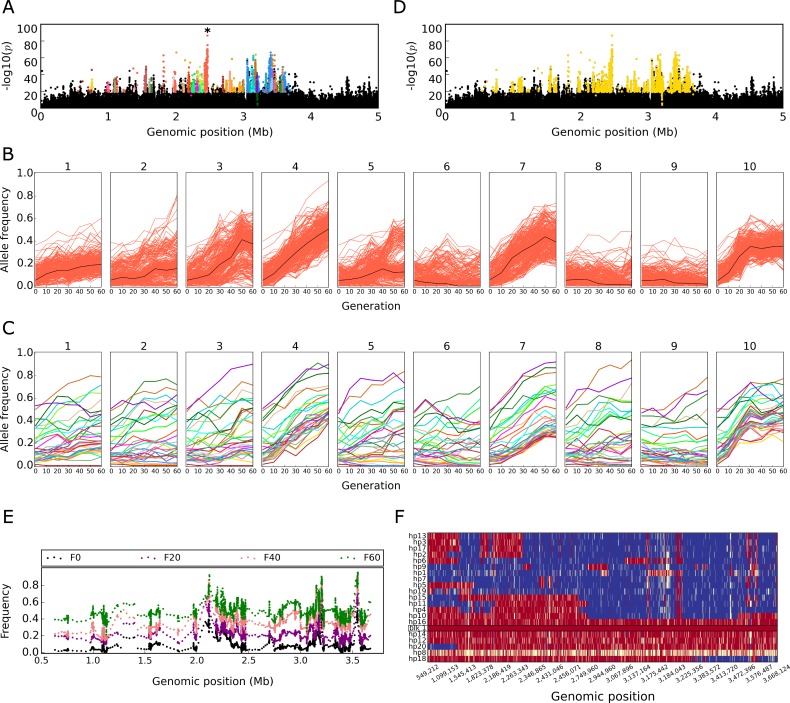
Fig 1. Reconstruction of selected haplotype block from Pool-Seq data.
Manhattan plots (panel A, D) show negative log10 transformed p-values from CMH test contrasting the ancestral (F0) with evolved (F60) populations (Chromosome 2L: 1–5 Mb). SNPs with highly correlated allele frequencies across replicates and time points are clustered together with stringent clustering (panel A), and each cluster is indicated by a different color. SNPs in each cluster (e.g., the orange cluster marked with an asterisk in panel A) have correlated frequency trajectories across 60 generations in 10 replicates (panel B); the black line depicts the median allele frequency trajectory. (C) Trajectories of the median allele frequency of the correlated SNPs are shown for each of the clusters in this region (color code corresponds to panel A). Despite different starting frequencies, the median trajectories greatly resemble each other, suggesting that they are correlated and reflect a large selected genomic region. (D) Combining the adjacent short clusters with less stringent correlation identifies a haplotype block with weakly correlated SNPs. All SNPs, which cluster together, are used as markers for this selected haplotype block. (E) Time-resolved allele frequencies of marker SNPs for the haplotype block (panel D) are plotted along their genomic positions. Each dot indicates the mean frequency of five SNPs in overlapping windows (offset = 1 SNP). The time-resolved allele frequencies show a consistent increase across the entire haplotype block throughout the experiment. The data are from replicate 4. (F) The reconstructed haplotype block (panel D, E) is experimentally validated. Rows represent 20 phased haplotypes from evolved replicate 4. The reconstructed haplotype block is indicated by a black frame. Each column corresponds to a marker SNP, with red color indicating the character state of the haplotype block, blue the alternative allele, and yellow an unknown nucleotide. Data deposited in the Dryad Repository: https://doi.org/10.5061/dryad.rr137kn. CMH, Cochran-Mantel-Haenszel; SNP, single nucleotide polymorphism.