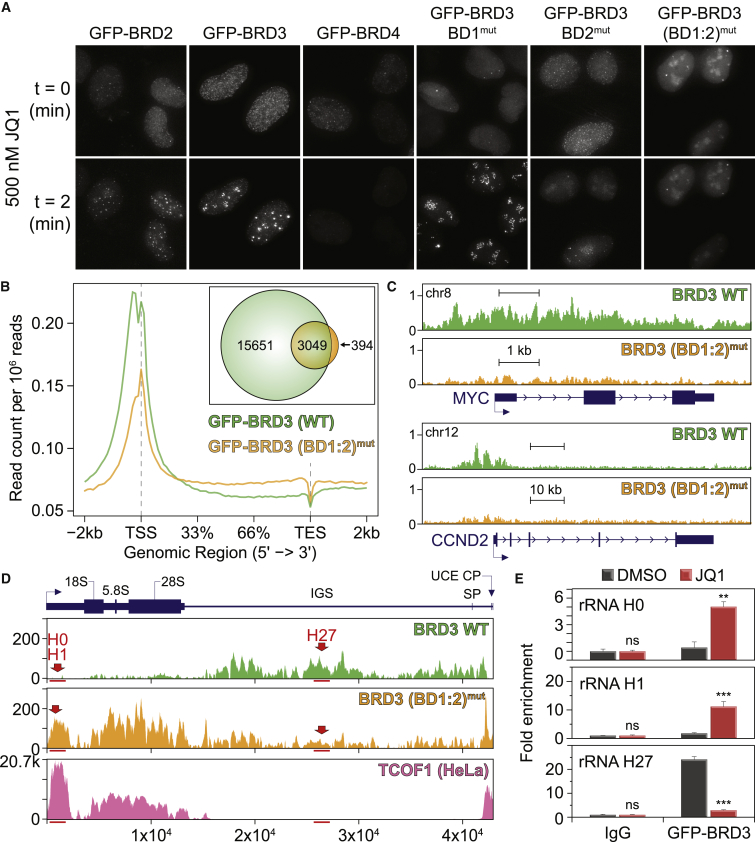
Figure 6.
BRD Inhibition Modulates BRD3 Localization
(A) Still images of indicated GFP-tagged BET constructs in live U-2 OS cells.
(B) Average BRD3 WT or (BD1:2)mut ChIP-seq read counts plotted over genes. TSS, transcription start site; TES, transcription end site. Inset: binding sites detected with each construct.
(C) Genome browser tracks showing BRD3 occupancy across the MYC and CCND2 gene loci. y axis: normalized read counts in reads per million per basepair.
(D) Schematic representation of a single human rDNA repeat relative to the transcription start site (TSS) of the rDNA repeat (x axis; based on GenBank U13369; SP: spacer promoter; UCE, upstream control element; IGS, intergenic spacer; CP, core promoter). y axis: normalized read counts of BRD3 WT, (BD1:2)mut, and TCOF1 (from HeLa cells; Calo et al., 2018).
(E) GFP-BRD3 ChIP-qPCR to rDNA H0, H1, and H27 (see D) with and without JQ1 for 1 hr. x axis: signal fold enrichment against rabbit IgG isotype control purifications. Data represent the mean ± SEM (n = 3) of two biological replicates. p values were calculated using Student’s t test and are represented by ∗∗∗p < 0.001; ∗∗p < 0.01; ns, not significant.