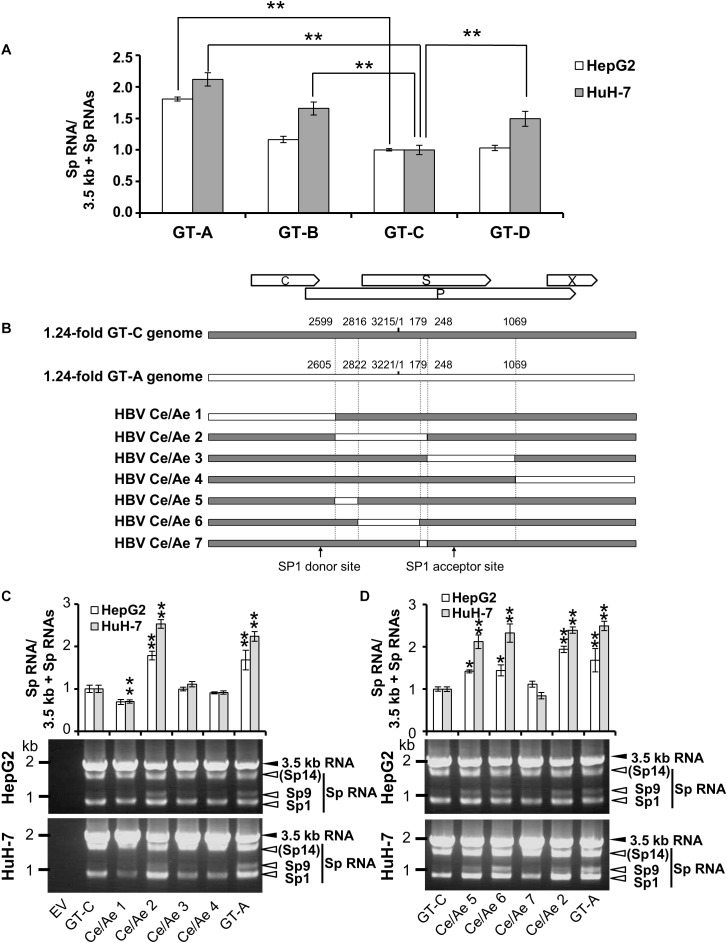
FIGURE 5.
Diversity in the splicing efficiency among HBV genotypes/strains. (A) pUC-HB-Ae (GT-A), -Bj56 (GT-B), -Ce (GT-C) or -D_Ind60 (GT-D) was transfected into HepG2 and HuH-7 cells. Two days after the transfection, total RNA was extracted from cells and analyzed by qRT-PCR to determine the levels of 3.5 kb RNA and spliced RNAs. The quantity ratios of the spliced RNAs to total 3.5 kb RNA derived RNA species were calculated and that of GT-C was set to 1. (B) A schematic representation of a series of mutated HBV genomes replacing parts of the GT-C sequence with corresponding parts of GT-A sequence. Positions of donor and acceptor sites of SP1 RNA are indicated. (C) qRT-PCR analysis was performed to determine 3.5 kb RNA and spliced RNA levels in HepG2 and HuH-7 cells transfected with pUC-HB- Ce (GT-C), -Ae (GT-A), HBV Ce/Ae1, /Ae2, /Ae3 or /Ae4. The quantity ratios of the spliced RNAs to total 3.5 kb RNA derived RNA species were calculated and those in GT-C-expressing cells were set to 1 (upper). Representative pattern for RT-PCR result indicating expression of unspliced and spliced (Sp) RNAs derived from 3.5 kb RNA in the transfected HepG2 (middle) and HuH-7 cells (lower). (D) As in (C) but mutated genomes HBV Ce/Ae5, /Ae6, /Ae7 as well as GT-C, -A and HBV Ce/Ae2 were used for expression of HBV RNAs. Values represent mean ± SD (HepG2; n = 2, HuH-7; n = 3). ∗p < 0.05, ∗∗p < 0.01, by one-way ANOVA followed by Dunnett’s test compared to GT-C.