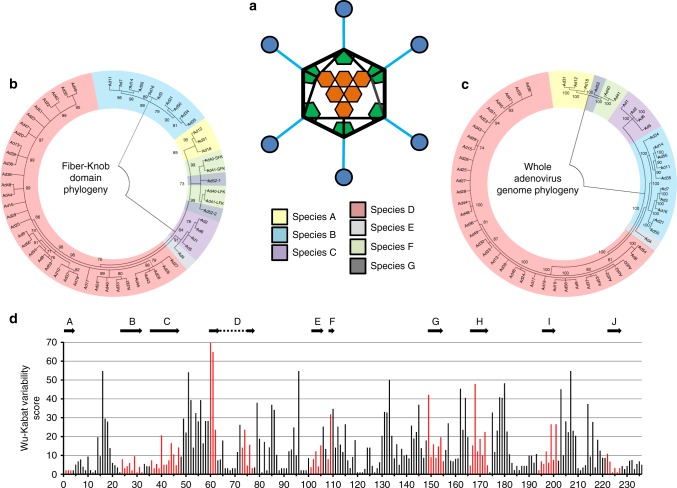
Fig. 1.
Phylogenetic analysis of adenoviruses mapped by whole genome and fiber-knob domain. A diagrammatic representation of the adenoviral major capsid proteins shows the icosahedral capsid structure with the Hexon (orange) comprising the facets, pentons (green) at the vertices, from which the fiber proteins (fiber-shaft in light blue, fiber-knob in dark blue) protrude (a). Condensed maximum likelihood trees (percentage confidence shown by numbers next to nodes) were generated from alignments of fiber-knob domain amino acid sequences of adenoviruses 1–56 (b) or whole genome NT sequences (c). Adenoviruses divide into 7 subspecies, as denoted in the key, regardless of alignment used, but the species D adenoviruses divide into additional sub-species when determined by fiber-knob alignment, for readability simple nomenclature is used, all are human adenovirus. Numbers next to nodes denote confidence. Wu–Kabat variability analysis of the Clustal Omega aligned fiber-knob domains amino acid sequences of adenoviruses 1–56 (d) reveals regions of low amino acid variability corresponding to beta-sheets. The locations of HAdV-C5 β-strands, as described by Xia et al. (1994)27, are aligned to the structure and shown by arrows, the corresponding positions are coloured in red