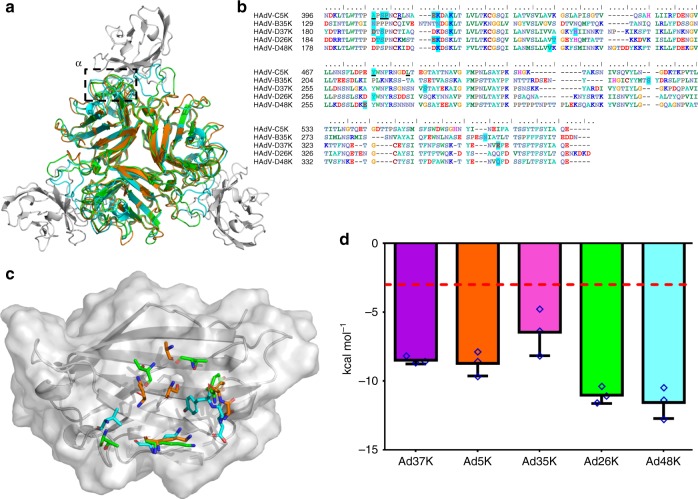
Fig. 5.
Modelling of the HAdV-D26K and HAdV-D48K interaction with CAR at the α-interface. The α-interface region is shown by the box on the structural alignment of HAdV-C5K (orange), 26K (green), and 48K (cyan) fiber-knob domain crystal structures in complex with CAR-D1 domain (grey) as determined by homology alignment to the previously reported Ad37K CAR-D1 structure (PDB: 2J12) (a). The aligned amino acid sequence of the investigated fiber-knobs (b) and the predicted α-interface forming CAR-D1 binding residues are highlighted in blue, with the underlined residues representing the HAdV-C5K amino acids shown by Kirby et al. (2000)37 to be important for CAR interaction. Conservation of key residues can be seen between HAdV-C5K, HAdV-D26K, and HAdV-D48K fiber-knobs. This conservation is visualised, with the contact residues comprising the α-interface with HAdV-C5K, 26K, and 48K shown as sticks in complex with the energy minimised CAR-D1 domain (grey), shown as the surface of the maximum spatial occupancy of the aligned CAR-D1 monomers from each of the energy minimised models in complex with HAdV-C5K, 26K, and 48K fiber-knobs (c). d Plots the predicted binding energy of the energy minimised fiber-knob proteins to CAR-D1 complex in the α-interface, only. Lower binding energy indicates a more stable interface with the red line depicting 3.0 kcal mol−1, which can be considered background. n = 3, where each calculation is an independent fiber-knob: CAR interface, error bars indicate mean ± SD