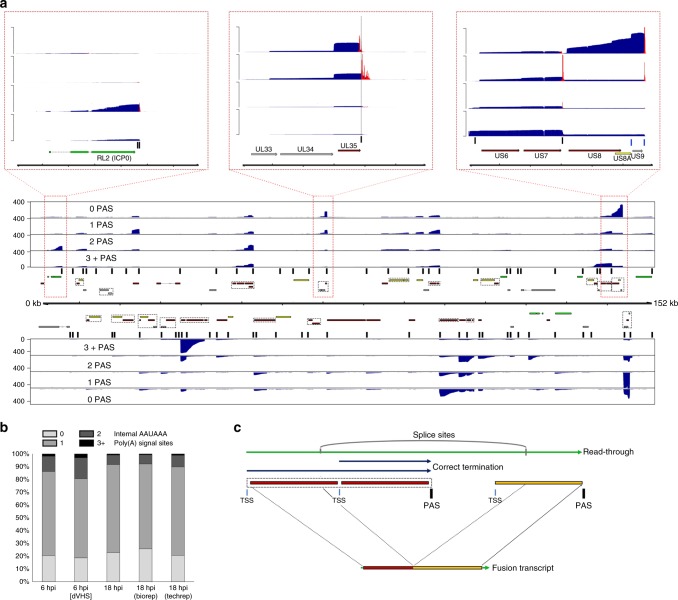
Fig. 5.
Detection of read-through transcription from the HSV-1 genome. a HSV-1 sequence reads were segregated according to the number of AAUAAA PAS motifs present and aligned against the HSV-1 genome to produce coverage plots showing the location of mapped reads. Here, complex (multiple overlapping ORFs) gene arrays are identified by cross-hatched boxes, while the locations of AAUAAA motifs are indicated by vertical black bars. The three inset boxes correspond to the red cross-hatched areas of the genome that exemplify (left) the presence of two AAUAAA motifs at the 3′ end of the RL2 transcript, (center) the position of pTTS sites relative to AAUAAA motifs (black vertical line), and (right) usage of the non-canonical AUUAAA motif (blue vertical line). pTTS estimates are shown as red overlays on the inset coverage plots. b HSV-1 transcription termination is generally initiated by recognition of a canonical (AAUAAA—dark gray) PAS sequence. Evidence of read-through transcription includes the presence of multiple AAUAAA motifs within a transcript and is observed in a small proportion 1–3% of HSV-1 mapping direct RNA-seq reads. c Transcription of HSV-1 genes initiates at transcription start sites (blue vertical line) and typically terminates shortly after traversing a canonical (AAUAAA) PAS sites (black vertical line). In rarer cases, termination does not occur and transcription extends further downstream as read-through until another PAS site is used. These extended transcripts may be subject to internal splicing which can give rise to fusion ORFs