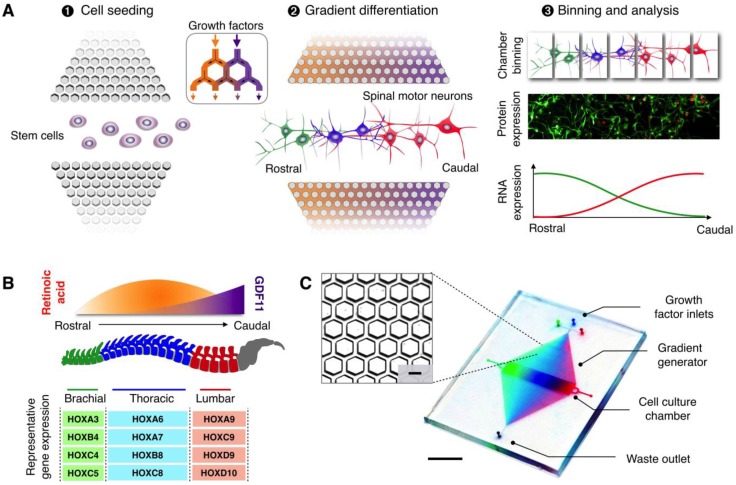
Figure 1.
MicroHIVE platform for directed differentiation of spinal motor neurons. (A) The microHIVE platform consists of a divergent honeycomb lattice of microhexagon structures that opens into a cell culture chamber, to enable stem cell seeding, generation of molecular gradients for directed cellular differentiation, and device binning and analysis. Seeded stem cells are exposed to fine spatiotemporal release of growth factors, whose flow streams branch and mix between the microhexagons to generate complex and high-resolution molecular gradients. These local molecular changes direct stem cell differentiation into an organized, continuous assembly of different spinal motor neuron subtypes. Through binning of the all-polymer culture chamber, the motor neuron assembly can be spatially dissected and its cellular identities characterized through RNA and protein measurements. (B) Schematic representation of an optimized molecular profile of retinoic acid and GDF11 to guide rostral-caudal motor neuron differentiation. The differentiated motor neurons can be categorized according to their associated regions in the spinal cord (i.e., brachial, thoracic and lumbar), as characterized by their differential expressions of region-associated HOX genes. (C) Photograph of the developed microHIVE platform. Scale bar indicates 1 cm. Insert shows a magnified view of the interlocking array of microhexagons. Scale bar of the insert indicates 100 μm.