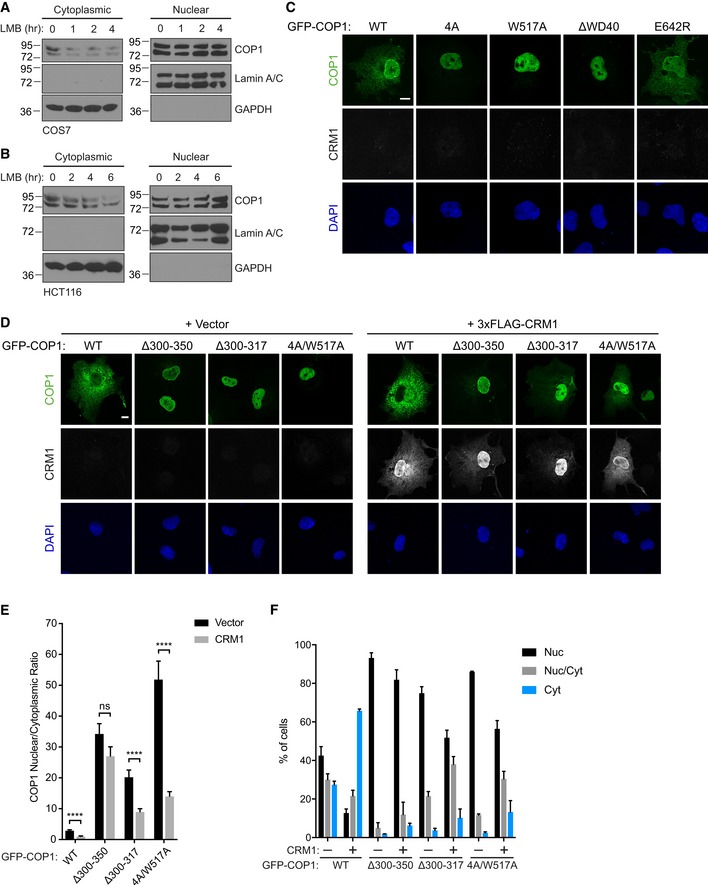
-
A, B
Subcellular fractionation of COS7 cells (A) and HCT116 cells (B) treated with 20 nM leptomycin B (LMB) for the indicated times, showing a reduction of endogenous COP1 levels in the cytoplasmic fraction over time.
-
C
Representative images of COS7 cells expressing the indicated GFP‐COP1 (green) constructs co‐transfected with empty vector. Cells were stained with anti‐FLAG (Sigma) and anti‐mouse Alexa Fluor 568. Nuclei were counterstained with DAPI (blue). Scale bar, 10 μm.
-
D
Representative images of COS7 cells expressing the indicated GFP‐COP1 (green) constructs co‐transfected with empty vector or 3xFLAG‐CRM1 (grayscale). Cells were stained with anti‐FLAG (Sigma) and anti‐mouse Alexa Fluor 568. Nuclei were counterstained with DAPI (blue). Scale bar, 10 μm.
-
E
Quantification of the average ratio of nuclear/cytoplasmic fluorescence of GFP‐COP1 constructs in (D). Mean values ± s.e.m. are shown for three independent experiments where 50 individual cells per experiment were analyzed. Significance was calculated using the Student's t‐test (ns, not significant; ****P < 0.00001).
-
F
Quantification of percentage of cells observed in (D) with the indicated GFP‐COP1 construct exhibiting nuclear (Nuc), nuclear and cytoplasmic (Nuc/Cyt), or cytoplasmic (Cyt) localization. Mean values ± s.e.m. are shown for three independent experiments where 50 individual cells per experiment were analyzed.