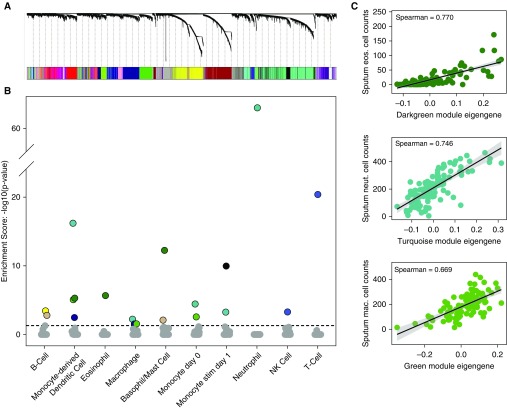
Figure 2.
Weighted gene coexpression analysis of sputum transcriptome data identifies gene networks reflective of specific immune cell types. (A) Weighted gene coexpression analysis hierarchical clustering dendrogram of the 13,536 genes that passed quality control filtering for induced sputum samples. Gene dissimilarity was calculated using topological overlap measure. The colors corresponding to network assignments are given in the bar below the dendrogram. (B) Plot of P values for gene set enrichment score of the immune cell–type gene sets in the sputum networks. Ten sputum networks were highly enriched with genes specific for different immune cells. For example, the dark green network was highly enriched with genes specific for eosinophils, basophils/mast cells, and dendritic cells. (C) Correlation plots between sputum cytospin cell counts and sputum network eigengene expression values. Specifically, correlations between the dark green module eigengene values and eosinophil cell counts, turquoise module eigengene values and neutrophil cell counts, and green module eigengene values and macrophage cell counts are shown. eos. = eosinophil; mac. = macrophage; neut. = neutrophil; NK = natural killer.