Fig. (1).
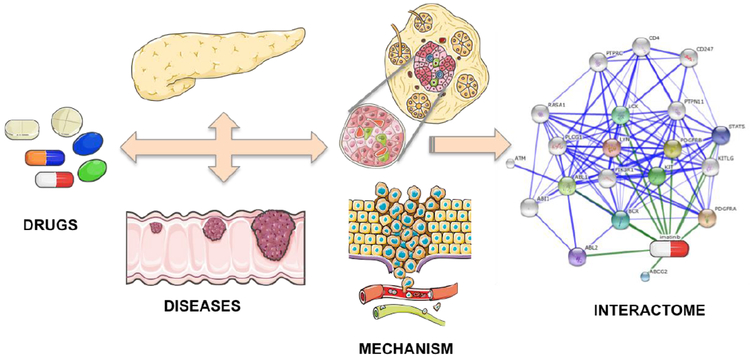
Interactome based therapeutic discovery paradigm used by the CANDO platform. Humans typically ingest a therapeutic drug (a small molecule or a biologic agent) with the goal of targeting particular indications (including infectious, inherited, and neoplastic diseases). The schematics for drug, disease and mechanism were adapted and modified from Servier Medical Art under Creative Commons CC-BY license. The visualization of the network of interaction between imatinib and its interactome was made using the STITCH4 platform [153]. In terms of mechanism, a given drug works by binding to one or more target proteins of interest, but also binds to other proteins causing off-target (neutral) and anti-target (negative) side effects. The CANDO platform makes interaction predictions for "all drugs" (currently a library of 3733 human approved compounds) against "all proteins" (currently a library of 48,278 proteins) to determine its efficacy against whole systems of interest, thereby enabling it to make putative drug predictions for “all” indications (that the library of drugs are currently approved for) simultaneously by performing comparative analyses of drug-proteome interaction signatures. In contrast to traditional drug discovery approaches focused on single targets, our innovative paradigm inverts the traditional one by first compiling a library of compounds/drugs that are safe to ingest with established side effects, and the shotgun virtual screen against whole proteomes/interactomes enables drug predictions for all indications simultaneously.
