Fig. (2).
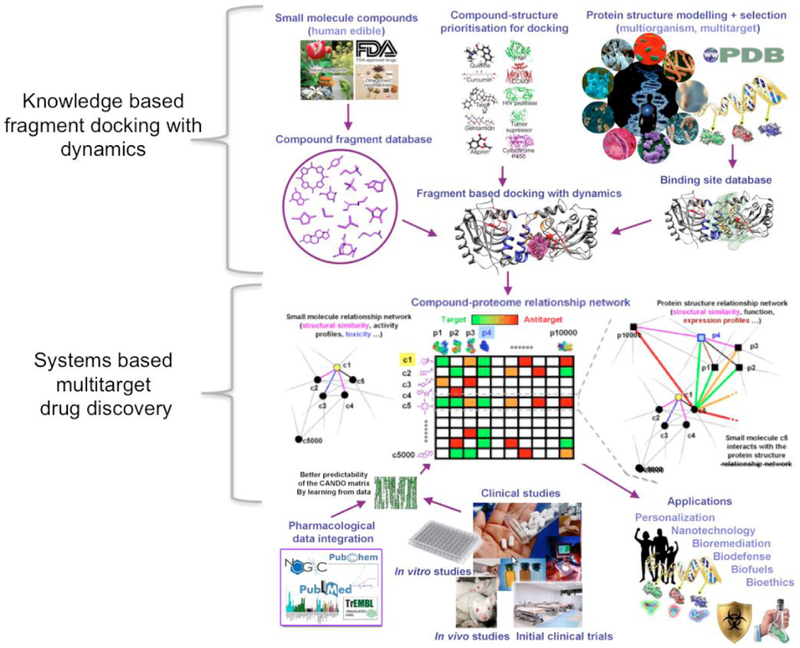
The Computational Analysis of Novel Drug Opportunities (CANDO) platform (http://protinfo.org/cando). The platform consists of a pipeline that generates the compound-proteome interaction matrix and indication-specific protocols to rank compounds that may be repurposed for particular indications/diseases given particular proteomes. The first version (v1) of the platform evaluates interactions between 3,733 human ingestible compounds that are associated with 2030 indications and 48,278 protein structures (46,784 of which are used in this study). Via a process of “contextualization”, where the predicted interactions are evaluated in the context of biomolecular data from a wide variety of sources including molecular docking simulation tools (including our knowledge-based fragment docking with dynamics algorithm), candidate interactions are ranked according to the degree of interaction and similarity for all indications. Predictions made using our systems-based multitarget drug discovery and repurposing platform are subjected to validation via in vitro binding, functional, and cellular assays, in vivo studies (if possible), and off-label use clinical studies. The resulting information is made available to a wide variety of users and applications via the web. The signature comparison and ranking approach used by the CANDO platform yielded benchmarking accuracies of 12-25% for 1439 indications with at least two approved compounds. 58/163 (35%) top ranking predictions had comparable or better activity relative to existing drugs in twelve prospective in vitro studies across ten indications, and represent novel repurposed therapies for indications such as dengue, dental caries, diabetes, herpes, lupus, malaria, and tuberculosis. Our approach can be tailored to specific meta genomes by modeling all the corresponding variant (mutant) protein structures and making predictions of personalized/precision drug regimens for particular individuals. Our blinded shotgun multitarget approach to drug discovery significantly enhances its efficiency, reduces drug development costs, and also provides a holistic framework for multiscale modeling of complex biological systems with broader applications in medicine and engineering.
