Abstract
BACKGROUND:
Metastasis is the primary cause of mortality in cancer patients. Therefore, elucidating the genetics and epigenetics of metastatic tumor cells and the mechanisms by which tumor cells acquire metastatic properties constitute significant challenges in cancer research.
OBJECTIVE:
To summarize the current understandings of the specific genotype and phenotype of the metastatic tumor cells.
METHOD and RESULT:
In-depth genetic analysis of tumor cells, especially with advances in the next-generation sequencing, have revealed insights of the genotypes of metastatic tumor cells. Also, studies have shown that the cancer stem cell (CSC) and epithelial to mesenchymal transition (EMT) phenotypes are associated with the metastatic cascade.
CONCLUSION:
In this review, we will discuss recent advances in the field by focusing on the genomic instability and phenotypic dynamics of metastatic tumor cells.
Keywords: metastasis, epithelial to mesenchymal transition (EMT), cancer stem cell, circulating tumor cells, cellular plasticity, phenotype dynamics
Background
The spread of tumor cells or “metastasis” has been considered the most advanced stage of tumor progression. Usually revealed as disease at multiple secondary sites, metastases cause the majority of cancer-related death in patients(Lambert et al., 2017). For metastasis to occur, a series of sequential steps must be achieved by tumor cells. At the primary site, the tumor cells proliferate, invade the surrounding stroma and enter the circulation directly through blood vessels or indirectly through the lymphatic system. The metastatic tumor cells will then be required to survive in the circulation, to adapt to the novel microenvironment of secondary organs and colonize these organs to become metastatic lesions. These overwhelming challenges constrain metastasis from being an efficient process(Luzzi et al., 1998; Valastyan and Weinberg, 2011; Lambert et al., 2017). Only a limited number of disseminated tumor cells are capable of forming secondary tumors. Metastatic inefficiency is determined by both intrinsic properties of tumor cells and extrinsic factors in the microenvironment. The intrinsic properties of tumor cells constitute the driving force for invasion into local tissue, survival in the circulation, and stem-like abilities to seed secondary tumors (Lambert et al., 2017; Liu and Cao, 2016). Moreover, specific intrinsic tumor-derived factors may also orchestrate the metastatic microenvironment by remodeling extracellular matrix, regulating angiogenesis, and recruiting immune cells in both the primary and metastatic organs (Liu and Cao, 2016). Therefore, in this review, we will focus on the intrinsic properties of tumor cells, including the genotypic and phenotypic features that define their metastatic abilities.
An unanswered question is whether metastatic tumor cells represent a unique subpopulation of the bulk primary tumor cells (Nguyen and Massague, 2007; Martinez-Cardus et al., 2016; Patel and Vanharanta 2016). It is known that primary tumor cells are heterogeneous at the genomic, epigenomic and phenotypic levels. Recent evidence regarding the complexity of intratumor heterogeneity suggested that multiple clones of cancer stem cells (CSCs) sharing the original oncogenic mutations may co-exist in the primary tumor. Each CSC subcolony individually accumulate unique mutations, allowing sequential selection of more aggressive subclones during tumor progression. Also, the differentiation of CSCs may be transient, bidirectional, and profoundly affected by microenvironmental factors and therapeutic regimens. The hypothesis of CSC plasticity and phenotypic dynamics fundamentally shapes our understanding of tumor progression.
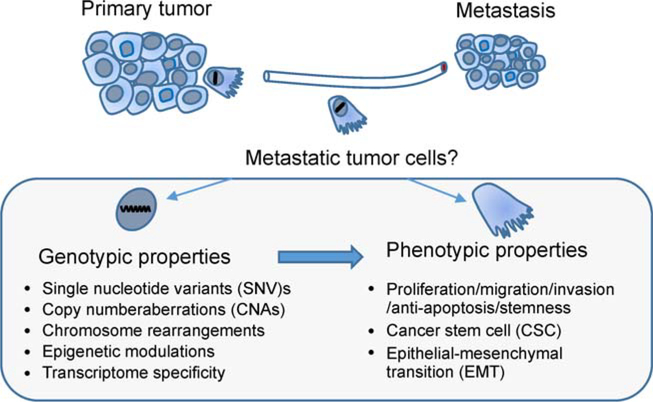
The identity of a metastatic tumor cell is still considered in terms of the CSC concept and clonal evolution theory. On the one hand, metastatic cells inherit critical driver mutations from their parental tumors allowing the persistence of tumorigenic ability. On the other hand, metastatic cells develop new mutations to adapt to emerging challenges during metastasis or treatment. It is imperative to determine the genetic, epigenetic and phenotypic characteristics that define metastatic tumor cells (Fig. 1); and whether such metastatic features are inherited or acquired transiently at a later stage during metastasis. More importantly, it is critical to determine whether these features are targetable for therapy.
Figure 1.
The genotypic and phenotypic properties that define the metastatic tumor cells. The specific features of metastatic tumor cells could be defined on genomic and phenotypic levels. Both germline and somatic alterations including SNVs, CNAs, chromosome rearrangement and epigenetic modulations would lead to changes in the transcriptome of metastatic tumor cells. These genomic changes would not only serve as evolution markers in metastasis, but also manifest functions of cell proliferation, migration, invasion, anti-apoptosis and stemness that would be closely related to the CSC and EMT phenotypes.
Genetics and genomics of metastatic tumor cells
In-depth genetic analysis of cancer cells has revealed cancer to be a complex genetic disease involving nearly 1,000 known cancer-related genes (~250 oncogenes, ~700 tumor suppressors)(Wishart, 2015). Alterations of these cancer-related genes include single nucleotide variants (SNVs), copy number alterations (CNAs), genomic rearrangements and epigenetic modulations. Of note, the genomic variations not only serve as markers of evolution but also support tumor progression by altering the expression levels and biological functions of the cancer-related genes. Particular genetic and genomic alterations may be specifically required for metastasis formation.
The genetic origins of a primary tumor could significantly impact its metastatic outcome. For example, using conditional mouse lung cancer models, expression of the activating KrasG12D mutation initiated tumors that remained localized in mice, whereas combination with Trp53 deletion triggered metastasis (DuPage et al., 2009). Similarly, in the prostate adenocarcinoma model initiated by Pten mutation, further loss of Rb1 leads to metastasis formation (Ku et al., 2017). Using a genome-wide association study (GWAS) approach, Zhu et al. (2017) found that expression of LIM-domain-only gene (LMO1) in a zebrafish model promoted metastatic neuroblastoma driven by the MYCN oncogene. Together, these studies suggest that cooperation of multiple genomic variations could promote the metastatic abilities of tumor cells. It is thereby not surprising that metastasis or recurrence may be predicted using the genomic hallmarks of the primary tumor. A comprehensive study of the molecular characteristics of pheochromocytomas and paragangliomas identified a unique pattern of somatically mutated driver genes including CSDE1, HRAS, RET EPAS1 and NF1 (Fishbein et al., 2017). Interestingly, a MAML3 fusion gene, which plays a role in Wnt signaling, was correlated with the metastatic phenotype in these rare tumor types, suggesting that metastasis-driving mutations could be tumor type-dependent.
To globally characterize the genetic alterations required for metastasis, the MSK-IMPACT project was initiated at Memorial Sloan Kettering Cancer Center (Zehir et al., 2017). This project involved the large-scale, prospective sequencing of cancer-related genes performed with specimens from more than 10 000 patients with advanced cancer (341 genes in 2 809 tumors and 410 genes in 8 136 tumors). In contrast to previous cancer deep-sequencing projects (e.g. the Cancer Genome Atlas (TCGA)) that were focused mainly on untreated primary cancers, the MSK-IMPACT cohort included patients receiving treatment before sequencing, with 43% of the specimens obtained from metastatic tumors. The MSK-IMPACT data revealed the consistently crucial roles of TP53, KRAS, PIK3CA, and BRAF in the metastatic tumors. Another recent whole-exome sequencing analysis of ~500 patients with metastatic tumors also discovered that TP53, CDKN2A, PTEN, PIK3CA, and RB1 were the most prevalent genes altered somatically in metastatic cancer (Robinson et al., 2017). In addition to somatic mutations, germline mutations in genes including BRCA1, BRCA2, CHEK2, and MUTYH, were also identified to be specifically associated with metastatic tumors. These mutated genes are commonly involved in DNA-repair pathways, indicating that genomic instability and the continuous accumulation of mutations may offer survival advantages to metastatic tumor cells.
The genetic features of metastatic tumors have also been studied by comparing serial biopsy specimens during tumor progression in the same patient (Campbell et al., 2010; Brastianos et al., 2015; Makohon-Moore and Iacobuzio-Donahue 2016; Yates et al., 2017). These studies have revealed a branched evolution of tumor cells, in which the primary tumors and metastases share a common ancestor, yet diverge to evolve independently later(Brastianos et al., 2015; Makohon-Moore and Iacobuzio-Donahue 2016). In pancreatic cancer patients, genomic rearrangements indicative of telomere dysfunction and abnormal cell-cycle control were demonstrated to occur predominantly in early cancer development and inherited by metastatic lesions (Campbell et al., 2010). Additional mutations that were absent in the primary tumors were identified in metastases, with an organ-specific pattern. By sequencing serial specimens of primary, locally relapsed, and metastatic breast cancers, it was recently showed that metastatic clones were disseminated late from primary tumors and continued to acquire mutations(Yates et al., 2017). The metastasis-specific mutations include clinically actionable alterations and mutations that inactivate the SWI/SNF and JAK2- STAT3 pathways. These results suggest that the metastases are genetically distinct from their parental tumors, although it is still not clear whether the additional mutations are prerequisites for metastasis.
In a recent study of pancreatic cancer metastases, it was shown that the same mutation in driver genes (genes that promote cancer development or progression) was shared between the primary and all metastatic lesions in the same patient (Makohon-Moore et al., 2017). Genomic analysis of multiple metastases in breast cancer patients also revealed similarities in overall signatures of gene expression, DNA copy numbers, and somatic mutations across the primary tumor and its associated metastases (Hoadley et al., 2016). The mutations that developed during metastasis were considered more likely to be passengers (mutations that do not affect cancer development or progression) with great individual variations and metastatic organ specificities. Of note, the definitions of the driver and passenger mutations could be challenging based on the frequency-based or function-based approaches to identify them (Pon and Marra, 2015). Especially for metastases, their genetic landscape may be fundamentally altered by choice of therapy. For example, 21.5% of metastatic breast cancer patients acquired CYP19A1 gene (encoding aromatase) amplification after aromatase inhibitor (AI) treatment, while such mutations only accounted for a minor fraction of those found in patients who had undergone another therapy (Magnani et al., 2017).
The transcriptome and its regulation of metastatic tumor cells
Although genomic studies have so far not been able to identify driver mutations specific for metastasis, metastatic tumor cells usually display remarkable specificity at the transcriptional level. By using a conditional lung cancer model (KrasG12D/Trp53−/−), tumor cells with the same driver mutations were isolated from distinct stages of metastatic progression (Chuang et al., 2017). Systematic genomic analysis of metastatic versus nonmetastatic tumor cells revealed a transcriptional program involving the CD109-Jak-Stat3 axis as a critical regulator of metastatic ability. Such transcriptome specificity of metastatic tumor cells suggests a special phenotypic rather than a genotypic requirement for metastasis formation.
Many genes strictly related to cell stemness and EMT phenotypes such as enhanced migration, invasion, anti-apoptosis, and self-renewal have been charaterized as metastasis-related genes. By isolating breast tumor cells that metastasize to different organs (e.g. lung, bone and brain), transcriptome signatures driving organ-specific metastasis were identified (Kang et al., 2003; Minn et al., 2005; Bos et al., 2009). However, there is very limited overlap among these metastatic signatures, indicating the particular requirements for metastasis formation in different organ microenvironments. Moreover, multiple signaling pathways may be required to cooperate at critical steps of metastasis. Transcriptional analysis of different subtypes of breast cancer revealed a specific correlation between the loss of expression of RasGAP tumor suppressor genes and aggressive luminal B breast tumors (Olsen et al., 2017). Functionally, these RasGAP genes cooperatively regulate RAS and NF-kB signaling that enhance metastatic features such as invasion and EMT, respectively.
Of note, the metastatic signatures were mostly identified by comparing the transcriptomes of non-metastatic and meta-static tumor cell lines, or primary tumors with different metastatic outcomes. However, paired analyses of the primary and established metastatic lesions typically revealed very similar transcriptomes (Brastianos et al., 2015; Yates et al., 2017), which may provide support for the dynamic phenotype theory. Metastatic features may be transiently gained by tumor cells during metastasis. As long as the metastatic tumor cells colonize the secondary organ, the cells could revert to their phenotypes of origin. Recently, using HMGA2 reporter mice, a transient subpopulation of pancreatic ductal adenocarcinoma (PDA) tumor cells (HMGA2 + ) was isolated with exceptionally high metastatic ability (Chiou et al., 2017). These metastatic cells highly expressed BLIMP1, a hypoxiainducible transcription factor, which was identified as a driver of PDA metastasis. Importantly, both HMGA2 and BLIMP1 were only expressed transiently in response to the hypoxic microenvironment of the primary tumor, and their expression was not detected in established metastatic lesions. These results provide evidence that a specific tumor microenvironment such as hypoxia may activate a dynamic metastatic phenotype of tumor cells.
Given the similar genetic alterations between primary and metastatic lesions, such transcriptome specificity of metastatic tumor cells is more likely to have been gained via epigenetic reprogramming. In a mouse model, FOXA1 transcription factor was implicated in promoting global enhancer activity in cells, and may play an essential role in the metastatic transition of PDA (Roe et al., 2017). An analysis of PDA patient samples has also demonstrated large-scale reprogramming of chromatin modifications during metastasis in the absence of specific driver mutations (McDonald et al., 2017). In particular, the distant metastases were found to have co-evolved a dependence on the oxidative branch of the pentose phosphate pathway (oxPPP), which suggested a model of metabolic-epigenetic programs in metastasis development. Also, DNA methylation seems to play a negative role in regulating tumor metastasis. Unique DNA hypomethylation patterns were observed at enhancers in Ewing sarcoma, due to the expression of the disease-defining EWS-FLI1 fusion protein (Sheffield et al., 2017). Interestingly, higher heterogeneity of intratumoral DNA methylation has been correlated to the metastatic status at diagnosis in Ewing sarcoma patients. Consistent with this, the deficiency of DNA methyltransferases, Dnmt3a and Dnmt3b, led to more aggressive and metastatic tumors in an epidermal carcinoma model (Rinaldi et al., 2017). Also, in patients with localized non-indolent prostate cancer, a combined genomic signature including ATM mutations, DNA methylation events, and MYC CNAs was highly discriminative of patients who would experience disease relapse (Fraser et al., 2017). This genomic signature even outperformed well-described prognostic biomarkers of prostate cancer.
Taken together, new advances in genomic and transcriptomic studies of metastatic tumors have revealed a complicated phylogenetic relationship with the primary tumors. The metastatic tumor usually shares the origin driver mutations and shows similar transcriptome profiles with the primary tumor. However, during the process of metastasis, tumor cells may accumulate additional passenger mutations due to genomic instability. Moreover, the overwhelming microenvironmental challenges could bestow the metastatic tumor cells specificity at the transcriptome level. These signatures of metastasis could be shaped by the different tumor types, organ-specific microenvironments and the anti-cancer treatments. Importantly, epigenetic re-programming plays a significant role in the transcriptional regulation of metastatic tumor cells. Such mechanisms may be versatile, reversible and dynamic in response to microenvironmental factors. These findings also support the concept that it is the genomic instability and phenotypic plasticity, but not the genotype or phenotype itself, that determines the eventual survival of metastatic tumor cells,
Phenotypes of metastatic tumor cells
Phenotypically, metastatic tumor cells are supposed to fit well in the concepts of “cancer stem cell” (CSC), which describes a cancer cell’s ability to self-renew and establish secondary tumors (Cabrera et al., 2015; Peitzsch et al., 2017), and the epithelial-to-mesenchymal transition (EMT), which represents a mechanism that confers migration/invasion abilities to cells (Mani et al., 2008; Brabletz 2012; Ye et al., 2015). Therefore, considerable efforts have been made to investigate the hypothesis that tumor cells with unique EMT/CSC signatures seed metastasis (Yu et al., 2013; Aceto et al., 2014; Lawson et al., 2015). However, no significant enrichment of CSCs or EMT tumor cells was observed in established metastatic lesions especially from clinic samples (Lim and Thiery, 2012; Chui, 2013;). Indeed, the stem and EMT features of tumor cells are concomitant with lower proliferation status, which represents a clear disadvantage for metastatic outgrowth (Lim and Thiery, 2012; Hecht et al., 2015). These findings indicate two possibilities: 1) metastatic cells dynamically change in phenotypes; 2) the CSC and EMT tumor cells may not be genuinely metastatic cells. In the following sections, we will discuss these discrepancies and controversies in interpreting the concepts of CSCs and EMT and their roles in metastasis.
Cancer stem cells and metastasis
CSCs were first described in acute myeloid leukemia as CD34+/CD38− cells, which resemble the normal hematopoietic stem cell phenotype (Lapidot et al., 1994). Since then, CSCs have been identified in solid tumors by different markers such as CD133, ALDH, OCT3/4, SOX2, PROCR, CD24, CD29, CD44, and NESTIN, (Cabrera et al., 2015; Nagare et al., 2017; Peitzsch et al., 2017). Notably, most of these markers are common to healthy adult stem cells. CSC markers may also be organ-specific, and there is a conspicuous lack of ubiquitous CSC markers for analyzing all tumor types.
In breast cancer, Al-Hajj et al., (Al-Hajj et al., 2003) first isolated CD44( + )CD24(-/low)Lineage(−) cells from human breast cancers, and showed that these cells had enhanced efficiency to form secondary tumors in mice. Subsequent studies showed that the CD44+ CSC population from both primary breast tumor and lung metastases were highly enriched for tumor-initiating cells, suggesting a direct contribution of CSCs to metastasis (Liu et al., 2010; Jaggupilli and Elkord, 2012). The identity and contributions of CSCs to tumor progression and metastasis have been investigated using numerous tumor models. A study using a TGF-beta receptor II gene (Tgfbr2)-deficient squamous cell carcinoma mouse model showed that ELMO1, a Rac-activating guanine exchange factor, was explicitly expressed in the CSCs in tumors at the transition zone (McCauley et al., 2017). Knocking down Elmo1 expression significantly impaired lung metastasis in the mouse model. Recently, in a colon cancer mouse model, Lgr5 (the leucine-rich repeat-containing G-protein-coupled receptor 5) was characterized as a reliable intestinal CSC marker (de Sousa e Melo et al., 2017). Of note, the Lgr5− population of tumor cells replenished Lgr5+ CSCs, and as a result, primary tumor regression was not observed upon specific ablation of Lgr5+ CSCs. Interestingly, the Lgr5+ CSCs were highly enriched in micro-metastatic lesions and were critical for metastasis formation. Depletion of Lgr5+ CSCs significantly impaired liver metastasis even in mice bearing established metastatic lesions. These results demonstrated a role for CSCs in metastasis, but also suggested that the specific organ microenvironment affected the requirement of CSCs for tumor maintenance.
Importantly, metastatic tumor cells may persist in a dynamic CSC state, in which they transition between a stem-cell and non-stem-cell state in response to microenvironmental factors. In long-term culture, CSCs and non-CSCs were able to repopulate each other, and both populations were capable of forming tumors (Gupta et al., 2009, Mani et al., 2008, Shackleton et al., 2009), suggesting that CSC marker expression could be transient and reversible. Indeed, a bivalent chromatin configuration in the promoter of ZEB1 (a key EMT regulator) plays an essential role in the switch between the CSC and non-CSC states in responding to microenvironmental signals (Chaffer et al., 2013). Since metastatic tumor cells are exposed to unpredictable challenges, their plasticity may better characterize metastatic cells than the presence of CSC markers.
Epithelial-to-mesenchymal transition and metastasis
EMT was initially observed in studies of embryo development, and described as a process whereby stationary epithelial cells transdifferentiate into motile mesenchymal cells (Thiery et al., 2009; Lim and Thiery, 2012). Through EMT, epithelial cells lose their tight connections with neighboring cells, exhibit resistance to apoptosis, and gain the ability to migrate and invade adjacent tissue. Also, the EMT program may bestow cancer cell stemness properties (Mani et al., 2008; Kalluri and Weinberg, 2009; Ye et al., 2015), thereby resembling the CSC phenotype. Since these EMT-associated features coincide substantially with the requirements for metastatic tumor cells, EMT has been proposed as an essential phenotype of metastasis.
EMT dynamics:
Continuous activation of the EMT program was shown to exhibit an inhibitory effect on metastasis. Overexpression of Twist1 (an EMT transcription factor) did not lead to outgrowth of macrometastases, although it promoted invasive primary tumor growth in a skin tumor model (Tsai et al., 2012). Instead, suppression of Twist1 expression was necessary for metastasis formation (Tsai et al., 2012). Similarly, SNAI1 (another EMT transcription factor) was found to be transiently expressed during breast cancer metastasis. Continuous SNAIL1 expression led to a significant decrease in number of lung metastases (Tran et al., 2014). The requirement for reversible expression of these EMT-promoting transcription factors supports a hypothesis of transient EMT, which could explain the observation that metastatic lesions usually mirror the epithelial phenotype of primary tumors (Valastyan and Weinberg, 2011; Lambert et al., 2017). Using a fluorescent-labeled patient-derived xenograft (PDX) model of breast cancer, Lawson et al. (Lawson et al., 2015) compared the transcriptomes of single metastatic tumor cells from animals with low versus high metastatic burden. Interestingly, gene expression signatures of stemness, EMT, pro-survival and dormancy were found in metastatic tumor cells from tissue with low metastatic burden, while the cells from tissues with high metastatic burden displayed more differentiated luminal gene expression signatures. These findings also suggested that dynamic phenotypic changes in metastatic tumor cells occur during tumor progression.
The controversy regarding the requirement for EMT in metastasis:
Recent studies using EMT lineage tracing models have raised vigorous discussion about the requirement of the EMT phenotype in metastasis. To trace the transient EMT events and clarify their contribution to metastasis, mesenchymal-specific Cre-mediated fluorescent marker switchable systems were established in spontaneous breast tumor mouse models (Fischer et al., 2015). In these models, tumor cells that have undergone EMT displayed a permanent switch in fluorescent marker expression from RFP to GFP. As expected, primary tumors predominantly exhibited RFP + epithelial phenotypes. Surprisingly, lung metastases were also mainly derived from RFP+ epithelial cells, questioning the direct contribution of EMT in metastasis (Fischer et al., 2015). One possible explanation was that the EMT lineage tracing models failed to report a transient EMT process (Ye et al., 2017). However, consistent EMT events were detected in the primary tumor, in the circulation, and in early lung metastases (Fischer et al., 2015). These disseminated EMT tumor cells were surpassed by their epithelial counterparts during metastasis formation. Furthermore, EMT blockade by ectopic expression of the miR-200 family did not affect lung metastasis (Fischer et al., 2015), indicating that EMT phenotype was not a prerequisite for metastasis-initiating cells. Also, using genetically engineered mouse models, Zheng et al.(2015) showed that genetic knockout of Snail or Twist (encoding EMT-inducing transcription factors) in mice did not alter the emergence of invasive PDA, its dissemination or liver metastasis. Both these studies (Fischer et al., 2015; Zheng et al., 2015) suggested that EMT was dispensable for metastasis, and proved contradictory to the prevailing hypothesis that EMT was essential for metastasis. Indeed, new lineage tracing systems and carcinogenesis models need to be evaluated to determine the generality of these observations.
Partial EMT:
The observations that cancer cells may harbor a spectrum of hybrid epithelial and mesenchymal states have led to the concept of partial EMT (Grigore et al., 2016; Yeung and Yang 2017). Unlike terminally differentiated epithelial cells, tumor cells have usually been observed to dedifferentiate with varying degrees of EMT phenotype (Kalluri and Weinberg, 2009). A spectrum of EMT status also exists among the tumor cells within an individual tumor. Using single-cell sequencing technology, the transcriptomes of hundreds of tumor cells from glioblastoma patients were profiled (Patel et al., 2014). Interestingly, most tumor cells did not fit into the distinct epithelial or mesenchymal cell types proposed by classical EMT models. These transcriptional profiles of individual tumor cells displayed a spectrum of intermediate phenotypes. Also, by analyzing more than 2,000 head and neck squamous cell carcinoma (HNSCC) cells, activation of a partial EMT(p-EMT) program was observed at the tumor leading edge (Puram et al., 2017). Of note, the overall expression of epithelial markers was apparently maintained in all tumor cells. The p-EMT program predominantly included genes encoding extracellular matrix proteins. Many classical EMT transcription factors such as SNAI1, TWIST1/2, and ZEB1/2 were not involved. Also, sequential analysis of sorted p-EMThigh and p-EMTlow cells further demonstrated that the activation of the p-EMT program during metastasis likely represents a transient state, since the differential expression of the p-EMT signature was not detected by comparing tumor cells from the primary and metastatic lesions (Puram et al., 2017). However, there is still lack of criteria for the partial EMT definition. It is still an open question whether it represents a special type of EMT or a reflection of the plasticity of tumor epithelial cells.
EMT status quantification:
The controversy regarding the contribution of EMT in metastasis is partially due to a lack of a standard quantification system, which defines the threshold between epithelial and mesenchymal phenotypes. To address this issue, efforts have been made to develop algorithms for EMT status score by weighting the expression of EMT-related genes (including both epithelial and mesenchymal markers) (Tan et al., 2014; George et al., 2017). A spectrum of EMT status was found in various cancers. However, EMT status did not necessarily correlate with poorer survival of cancer patients. Intriguing results were observed wherein the mesenchymal phenotype was prognostic for worse survival in some cancer types and better survival in others. Interestingly, when investigating the correlation of the EMT score with metastasis potential in breast cancer patients (George et al., 2017), metastatic cancer was either categorized as epithelial or a hybrid phenotype, indicating that a complete EMT phenotype is unnecessary for metastasis.
However, the use of bulk tumor transcriptomes for EMT scoring analysis has the complication that variations may derive from the transcriptomes of different stromal components rather than the EMT status of tumor cells (Goossens et al., 2015). Advances in technologies such as single cell DNA and RNA sequencing may provide critical connections between genotypes and phenotypes (Navin et al., 2011; Navin 2015), offering new perspectives on the translational applications of EMT in prognosis and personalized therapy.
Taken together, the evolving concepts of dynamic EMT and partial EMT have spurred further research to yield a quantitative metric to define such hybrid states, so that their functional role in metastasis can be investigated. Future studies employing sensitive and multiple EMT reporters have the potential to address some of the conflicting findings in the EMT field.
CSC/EMT phenotypes in Circulating tumor cells:
En route to metastasis, tumor cells travel through the bloodstream as Circulating tumor cells (CTCs) to reach distal organs, thus the phenotype of CTCs may reflect metastatic features. The traditional technique of CTC isolation (using the CellSearch system) depending on an epithelial marker has been challenged due to the existence of CTCs with a mesenchymal phenotype. Using a cocktail of antibodies against epithelial marker EpCam, as well as EGFR, and HER2, Yu et al. (2013) captured CTCs using a microfluidic herringbone-chip. CTCs with both epithelial (keratins, EpCam, and E-cadherin) and mesenchymal (fibronectin, N-cadherin, and PAI1) markers were identified. Interestingly, CTCs with mesenchymal phenotypes were enriched particularly in patients with triple negative breast cancer and associated with metastasis development (Yu et al., 2013). Also, stem-like markers (CD44+/CD24−) have been used in isolating CTCs in breast cancer patients. It was clear that the enumeration of CTCs itself seems not adequate to predict the metastasis outcomes, while the phenotype of CTCs such as re-proliferation ability showed correlation with the presence of brain metastasis in patients (Boral et al., 2017).
Recently, attractive techniques were developed to distinguish CTC phenotypes on single cell level by nanoparticle-mediated magnetic ranking (Poudineh et al., 2017). Using nanoparticles coated with anti-EpCam antibody, CTCs in the whole blood could be recovered according to their EpCam expression levels in different magnetic zones. Similarly, using RNA probes that are conjugated to the magnetic nanoparticles, a quantification analysis of interested gene expression in CTCs could be achieved with high sensitivity and efficacy (Labib et al., 2018). In addition, novel technologies for analyzing the proteomics of CTCs, especially for the phosphorylation and proteolytic processing, were also under development (Sinkala et al., 2017). These techniques hold the potentials to reveal the dynamic phenotypes of CTCs that are required for the metastasis-seeding.
Therapeutic targeting of the CSC/EMT phenotypes
Study of the CSC/EMT phenotype in metastasis formation has been driven by the prospects of developing targeted therapies for metastasis. However, the complexities of the plasticity and dynamics of these phenotypes predict that targeting therapies may only provide temporary effects, since new CSC or EMT tumor cells may continue to arise from the remaining tumor cell population that has not been targeted (Davis et al., 2014; Dragu et al., 2015). Also, complications occur due to the potentially reversible nature of the EMT process (Davis et al., 2014). Inhibiting EMT may impair the invasion and dissemination of tumor cells at the primary site, but may also promote cell proliferation and colonization at the metastatic site. This strategy would be unacceptable for patients, whose tumor cells may have already disseminated at the time of diagnosis.
Nevertheless, platforms that enable the discovery of specific cytotoxic agents or inhibitors of CSC/EMT tumor cells have been investigated. Using genetically modified human breast epithelial cells mimicking the CSC/EMT status of cancer cells, high-throughput screening for selective inhibitors of CSC growth has been investigated (Gupta et al., 2009; Carmody et al., 2010). Also, TGFβ, a major EMT-inducing factor, has been intensively studied as an anti-EMT target. Clinical trials using LY2157299, a TGFβRI inhibitor, either as monotherapy or in combination with standard chemotherapies, are ongoing in glioblastoma, pancreatic cancer and liver cancer patients (Giannelli et al., 2014; Rodon et al., 2015).
In addition, the association of CSC/EMT phenotype and chemoresistant properties provided another rationale for targeting the CSC/EMT tumor cells, which may achieve a synergistic effect with traditional chemotherapy. Indeed, inhibiting the EMT process by expression of the miR-200 family led to significant reduction in chemoresistant metastasis formation in a breast tumor model (Fischer et al., 2015). Also, in a model of squamous cell carcinoma, genetic ablation of BMI1+ CSCs significantly inhibits lymph metastasis development, and inhibition of BMI1 sensitized the tumor to cisplatin treatment (Chen et al., 2017). Therefore, the combination of anti-EMT/anti-CSC approaches with traditional chemotherapy may provide potential regimens to overcome chemoresistance and metastasis development.
Conclusions and perspectives
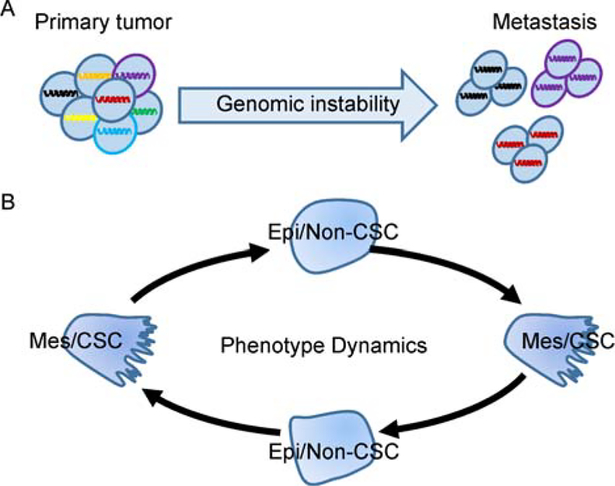
After years of searching for specific genetic, genomic, epigenetic and phenotypic features of metastatic tumor cells, it remains inconclusive whether metastatic tumor cells are as a special subpopulation within the bulk primary tumor cells. Although many genotypic and phenotypic markers specified for metastatic tumor cells have been proposed, these markers are vulnerable and cancer type dependent. Instead, the current findings suggest that metastases arise from multiple subclones of the primary tumor and share critical driver mutations (Fig. 2A). The genomic instability program of tumor cells leads to additional genomic alterations. However, such modifications are usually passenger mutations and are profoundly affected by the microenvironmental factors and treatments encountered during metastasis.
Figure 2.
The genotypic instability and phenotype dynamics of the metastatic tumor cells. A, Metastases could be derived from multiple subclones of the primary tumor cells. Genomic studies reveal a complicated phylogenetic relationship between the primary tumor and metastasis. Similar driving mutations and genetic alterations are shared between them, while additional passenger mutations are also detected in the metastases due to the genomic instability feature. B, The phenotype features of metastatic tumor cells could be transient, reversible between mesenchymal/stem like (Mes/CSC) and epithelial/non-stem like (Epi/non-CSC), and highly affected by microenvironment factors. It might not be the genotype or phenotype itself, but the genomic instability and phenotypic plasticity that determines the eventual survival of metastatic tumor cells.
Phenotypically, metastatic tumor cells require stemness to rebuild the hierarchy of tumor cell populations in metastatic lesions. However, no consistent CSC marker has been identified among tumors, yet CSCs and non-CSCs can repopulate each other under specific conditions. Tumor cells may exhibit a reversible EMT process during metastasis, and there is a spectrum of intermediate phenotypes (Fig. 2B). Given that metastatic tumor cells encounter very different microenvironments in the primary tumor, in the circulation, and in secondary organs, either the CSC or the EMT phenotype could provide a survival or proliferative advantage. These findings also suggest that future metastasis studies should focus on factors controlling cellular plasticity of metastatic tumor cells rather than on the phenotype itself.
Unfortunately, defining, tracking or measuring the phenotypic plasticity of metastatic tumor cells is challenging, particularly in vivo. Important questions still remain unanswered, such as: 1) Are there genetic or epigenetic markers of cellular plasticity? 2) How do cells control their plasticity? 3) What are the crucial mediators of cellular plasticity? And more importantly, 4) is phenotypic plasticity targetable to achieve therapeutic benefit in patients with metastatic disease? Equipped with much advanced techniques including single-cell sequencing, in vivo imaging, genetic engineered mouse and patient-derived xenograft (PDX) models, we are poised to unravel the temporal/spatial mysteries of metastatic tumor cells. Clarification of the (epi)genotypes underlying the phenotypic behavior of metastatic tumor cells will take our understanding of metastasis further and should culminate in successful therapeutic strategies against metastasis.
Footnotes
Compliance with ethics guidelines
Authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H(2014). Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell, 158:1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003). Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A, 100:3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boral D, Vishnoi M, Liu HN, Yin W, Sprouse ML, Scamardo A, Hong DS, Tan TZ, Thiery JP, Chang JC (2017). Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat Commun, 8:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos PD, Zhang XHF, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, Van de Vijver M, Gerald W, Foekens JA, Massagué J( 2009). Genes that mediate breast cancer metastasis to the brain. Nature, 459:1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T 2012. To differentiate or not–routes towards metastasis In: Nat Rev Cancer. England: p. 425–436. [DOI] [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K (2015). Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov, 5:1164–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera MC, Hollingsworth RE, Hurt EM (2015). Cancer stem cell plasticity and tumor hierarchy. World J Stem Cells, 7:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML (2010). The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature, 467:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody L, Germain A, Morgan B, VerPlank L, Fernandez C, Forbeck E, Ting A, Feng Y, Perez J, Dandapani S (2010). Identification of a Selective Small-Molecule Inhibitor of Breast Cancer Stem Cells-Probe 1 In: Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): National Center for Biotechnology Information (US). [PubMed] [Google Scholar]
- Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA, Weinberg RA (2013). Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell, 154:61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wu M, Li Y, Chang I, Yuan Q, Ekimyan-Salvo M, Deng P, Yu B, Yu Y, Dong J (2017). Targeting BMI1(+) Cancer Stem Cells Overcomes Chemoresistance and Inhibits Metastases in Squamous Cell Carcinoma. Cell Stem Cell, 20:621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou SH, Risca VI, Wang GX, Yang D, Gruner BM, Kathiria AS, Ma RK, Vaka D, Chu P, Kozak M (2017). BLIMP1 Induces Transient Metastatic Heterogeneity in Pancreatic Cancer. Cancer Discov, 7:1184–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Greenside PG, Rogers ZN, Brady JJ, Yang D, Ma RK, Caswell DR, Chiou SH, Winters AF, Gruner BM (2017). Molecular definition of a metastatic lung cancer state reveals a targetable CD109-Janus kinase-Stat axis. Nat Med, 23:291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui MH (2013). Insights into cancer metastasis from a clinicopathologic perspective: Epithelial-Mesenchymal Transition is not a necessary step. Int J Cancer, 132:1487–1495 [DOI] [PubMed] [Google Scholar]
- Davis FM, Stewart TA, Thompson EW, Monteith GR (2014). Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharmacol Sci, 35: 479–488 [DOI] [PubMed] [Google Scholar]
- de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H (2017). A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature, 543:676–680 [DOI] [PubMed] [Google Scholar]
- Dragu DL, Necula LG, Bleotu C, Diaconu CC, Chivu-Economescu M (2015). Therapies targeting cancer stem cells: Current trends and future challenges. World J Stem Cells, 7:1185–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T (2009). Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc, 4(7): 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J (2015). Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. November 26;527:472–476. Epub 2015/November/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, Lichtenberg TM, Murray BA, Ghayee HK, Else T(2017). Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell, 31:181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah YJ, Yousif F, Lin X, Masella AP(2017). Genomic hallmarks of localized, non-indolent prostate cancer. Nature, 541: 359–364 [DOI] [PubMed] [Google Scholar]
- George JT, Jolly MK, Xu S, Somarelli JA, Levine H (2017). Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res, 77: 6415–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannelli G, Villa E, Lahn M (2014). Transforming growth factor-beta as a therapeutic target in hepatocellular carcinoma. Cancer Res, 74:1890–1894 [DOI] [PubMed] [Google Scholar]
- Goossens N, Hoshida Y, Aguirre-Ghiso JA(2015). Origin and interpretation of cancer transcriptome profiling: the essential role of the stroma in determining prognosis and drug resistance. EMBO Mol Med, 7:1385–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigore AD, Jolly MK, Jia D, Farach-Carson MC, Levine H (2016). Tumor Budding: The Name is EMT. Partial EMT. J Clin Med, 29:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Chaffer CL, Weinberg RA (2009). Cancer stem cells: mirage or reality? Nat Med, 15:1010–1012 [DOI] [PubMed] [Google Scholar]
- Gupta, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES (2009). Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell, 138:645–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht I, Natan S, Zaritsky A, Levine H, Tsarfaty I, Ben-Jacob E(2015). The motility-proliferation-metabolism interplay during metastatic invasion. Sci Rep, 5:13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Siegel MB, Kanchi KL, Miller CA, Ding L, Zhao W, He X, Parker JS, Wendl MC, Fulton RS(2016). Tumor Evolution in Two Patients with Basal-like Breast Cancer: A Retrospective Genomics Study of Multiple Metastases. PLoS Med, e1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggupilli A, Elkord E (2012). Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin Dev Immunol, 2012: 708036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA (2009). The basics of epithelial-mesenchymal transition. J Clin Invest, 119:1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J (2003). A multigenic program mediating breast cancer metastasis to bone. Cancer Cell, 3:537–549 [DOI] [PubMed] [Google Scholar]
- Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Gomez EC, Wang J, Long HW, Xu B, Brown M, Loda M, Sawyers CL, Ellis L, Goodrich DW (2017). Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science, 355:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib M, Mohamadi RM, Poudineh M, Ahmed SU, Ivanov I, Huang CL, Moosavi M, Sargent EH, Kelley SO (2018). Single-cell mRNA cytometry via sequence-specific nanoparticle clustering and trapping. Nat Chem, 10:489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AW, Pattabiraman DR, Weinberg RA (2017). Emerging Biological Principles of Metastasis. Cell, 168:670–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature, 367:645–648 [DOI] [PubMed] [Google Scholar]
- Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, Yaswen P, Goga A, Werb Z (2015). Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature, 526:131–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Thiery JP 2012. Epithelial-mesenchymal transitions: insights from development In: Development. England: p. 3471–3486. [DOI] [PubMed] [Google Scholar]
- Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, Wen S, Chang YF, Bachmann MH, Shimono Y, Dalerba P, Adorno M, Lobo N, Bueno J, Dirbas FM, Goswami S, Somlo G, Condeelis J, Contag CH, Gambhir SS, Clarke MF (2010). Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A, 107:18115–18120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao X (2016). Characteristics and Significance of the Premetastatic Niche. Cancer Cell, 30:668–681 [DOI] [PubMed] [Google Scholar]
- Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, Groom AC (1998). Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol. 153:865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L, Frige G, Gadaleta RM, Corleone G, Fabris S, Kempe H, Verschure PJ, Barozzi I, Vircillo V, Hong SP, Perone Y, Saini M, Trumpp A, Viale G, Neri A, Ali S, Colleoni MA, Pruneri G, Minucci S (2017). Acquired CYP19A1 amplification is an early specific mechanism of aromatase inhibitor resistance in ERalpha metastatic breast cancer. Nat Genet,49:444–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makohon-Moore A, Iacobuzio-Donahue CA (2016). Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer,16:553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, Chatterjee K, Wong F, Jiao Y, Kohutek ZA, Hong J, Attiyeh M, Javier B, Wood LD, Hruban RH, Nowak MA, Papadopoulos N, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA (2017). Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet, 49:358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA (2008). The epithelialmesenchymal transition generates cells with properties of stem cells. Cell, 133:704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cardus A, Moran S, Musulen E, Moutinho C, Manzano JL, Martinez-Balibrea E, Tierno M, Elez E, Landolfi S, Lorden P, Arribas C, Müller F, Bock C, Tabernero J, Esteller M (2016). Epigenetic homogeneity within colorectal tumors predicts shorter relapse-free and overall survival times for patients with locoregional cancer. Gastroenterology, 151:961–972 [DOI] [PubMed] [Google Scholar]
- McCauley HA, Chevrier V, Birnbaum D, Guasch G (2017). De-repression of the RAC activator ELMO1 in cancer stem cells drives progression of TGFbeta-deficient squamous cell carcinoma from transition zones. Elife, 21:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S, Stauffer KM, Makohon-Moore A, Zhong Y, Wu H, Wellen KE, Locasale JW, Iacobuzio-Donahue CA, Feinberg AP (2017). Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet, 49:367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J ( 2005). Genes that mediate breast cancer metastasis to lung. Nature, 436:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagare RP, Sneha S, Priya SK, Ganesan TS (2017). Cancer Stem Cells-Are Surface Markers Alone Sufficient? Curr Stem Cell Res Ther, 12 (1): 37–44 [DOI] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M (2011). Tumour evolution inferred by single-cell sequencing. Nature, 472:90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin NE (2015). The first five years of single-cell cancer genomics and beyond. Genome Res, 25:1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Massague J (2007). Genetic determinants of cancer metastasis. Nat Rev Genet, 8:341–352 [DOI] [PubMed] [Google Scholar]
- Olsen SN, Wronski A, Castano Z, Dake B, Malone C, De Raedt T, Enos M, DeRose YS, Zhou W, Guerra S, Loda M, Welm A, Partridge AH, McAllister SS, Kuperwasser C, Cichowski K (2017). Loss of RasGAP tumor suppressors underlies the aggressive nature of luminal b breast cancers. Cancer Discov, 7:202–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science, 344:1396–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SA, Vanharanta S (2016). Epigenetic determinants of metastasis. Mol Oncol, 11(1): 79–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitzsch C, Tyutyunnykova A, Pantel K, Dubrovska A (2017). Cancer stem cells: The root of tumor recurrence and metastases. Semin Cancer Biol, 44(February): 10–24 [DOI] [PubMed] [Google Scholar]
- Pon JR, Marra MA (2015). Driver and passenger mutations in cancer. Annu Rev Pathol, 10(1): 25–50 [DOI] [PubMed] [Google Scholar]
- Poudineh M, Aldridge PM, Ahmed S, Green BJ, Kermanshah L, Nguyen V, Tu C, Mohamadi RM, Nam RK, Hansen A (2017). Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat Nanotechnol, 12:274–281 [DOI] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS (2017). Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell, 171:1611–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L, Avgustinova A, Martin M, Datta D, Solanas G, Prats N, Benitah SA (2017). Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR-gamma. Elife, 20:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V (2017). Integrative clinical genomics of metastatic cancer. Nature, 548: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodon J, Carducci MA, Sepulveda-Sanchez JM, Azaro A, Calvo E, Seoane J, Brana I, Sicart E, Gueorguieva I, Cleverly AL(2015). First-in-human dose study of the novel transforming growth factor-beta receptor I kinase inhibitor LY2157299 monohydrate in patients with advanced cancer and glioma. Clin Cancer Res, 21:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, Maiorino L, Tiriac H, Young CM, Miyabayashi K (2017). Enhancer reprogramming promotes pancreatic cancer metastasis. Cell, 170:875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Quintana E, Fearon ER, Morrison SJ (2009). Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell, 138:822–829 [DOI] [PubMed] [Google Scholar]
- Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schonegger A, Schuster M, Hadler J, Surdez D, Guillemot D, Lapouble E (2017). DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med, 23:386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkala E, Sollier-Christen E, Renier C, Rosas-Canyelles E, Che J, Heirich K, Duncombe TA, Vlassakis J, Yamauchi KA, Huang H (2017) . Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun, 8:14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, Thiery JP (2014). Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med, 6:1279–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA (2009). Epithelial-mesenchymal transitions in development and disease. In: Cell. United States. p. 871–890. [DOI] [PubMed] [Google Scholar]
- Tran HD, Luitel K, Kim M, Zhang K, Longmore GD, Tran DD (2014). Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res, 74:6330–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J (2012). Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell, 22:725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell, 147:275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS (2015). Is Cancer a Genetic Disease or a Metabolic Disease? In: EBioMedicine. p. 478–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, Alexandrov LB, Van Loo P, Haugland HK, Lilleng PK, ( 2017). Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell, 32:169–184.e167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Brabletz T, Kang Y, Longmore GD, Nieto MA, Stanger BZ, Yang J, Weinberg RA (2017). Upholding a role for EMT in breast cancer metastasis. Nature, 547:E1–e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, Weinberg RA (2015). Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature, 525: 256–260. Epub 2015/September/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KT, Yang J. 2017. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol, 11:28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM (2013). Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science, 339:580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med, 23:703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R (2015). Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature, 527:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhang X, Weichert-Leahey N, Dong Z, Zhang C, Lopez G, Tao T, He S, Wood AC, Oldridge D (2017). LMO1 Synergizes with MYCN to Promote Neuroblastoma Initiation and Metastasis. Cancer Cell, 32:310–323.e315 [DOI] [PMC free article] [PubMed] [Google Scholar]