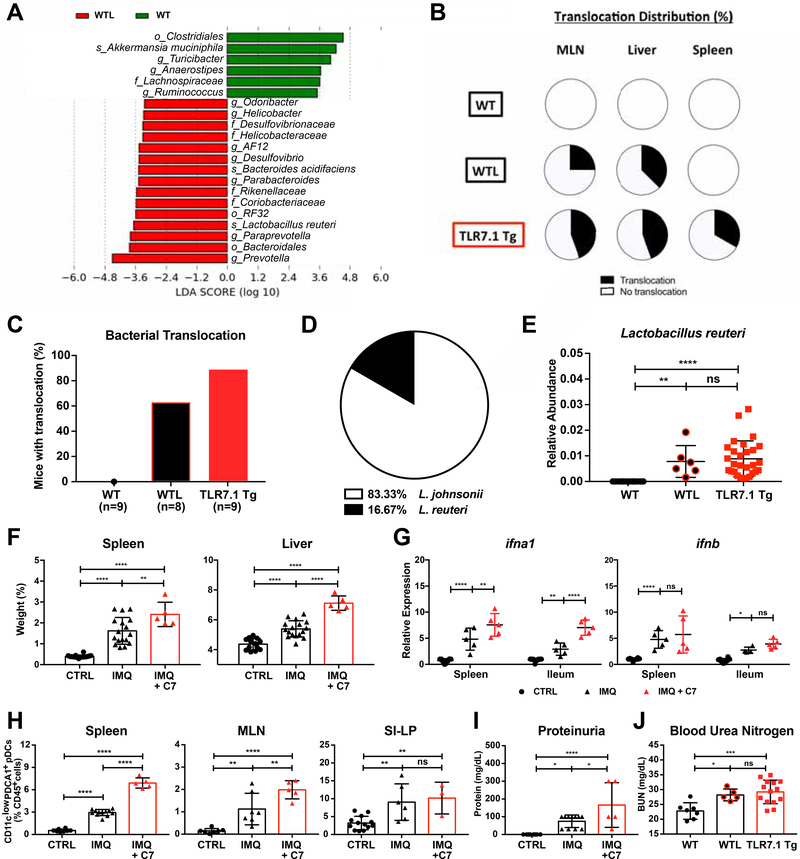
Figure 3. Bacterial communities increased in TLR7-dependent lupus models are transferable and exacerbate lupus-related pathogenesis.
Fecal pellets from WT mice and WT littermates (WTL) cohoused with TLR7.1 Tg were collected; bacterial DNA was isolated and sequenced. (A) LDA scores of altered taxa in WTL compared to WT. Shown are significantly altered taxa (increased in red, decreased in green). (B) Distribution of Lactobacillus spp. translocation in MLN, liver and spleen from WT, WTL, and TLR7.1 Tg mice. (C) Total percentage of bacterial translocation as determined by culture. (D) Profile of translocating bacterial species in WTL mice. (E) Relative abundances of fecal L. reuteri (LR) in WT, WTL and TLR7.1 Tg mice evaluated by 16S rDNA gene sequencing. (F-J) SPF C57B1/6 mice were treated with IMQ and gavaged with cecal microbial content from 12-week-old TLR7.1 Tg mice (C7). Mice were sacrificed for organ weights, immunologic, and histologic analyses. (F) Weights of spleen and liver. (G) Relative mRNA expression of type I IFN in spleen and ileum. Cells from spleen, MLN, and SI-LP were isolated and analyzed by flow cytometry. (H) Frequency of pDCs in spleen, MLN, and SI-LP. (I) Quantification of proteinuria. Q) Blood urea nitrogen levels in WTL. The results are expressed as mean ± SEM (n=5–20 mice per group). The results are representative of at least two independent experiments. *P<0.05 was considered statistically significant; **P<0.01; ***P<0.001; ****p<0.0001; ns=not significant. See also Figure S3 and Table S3.