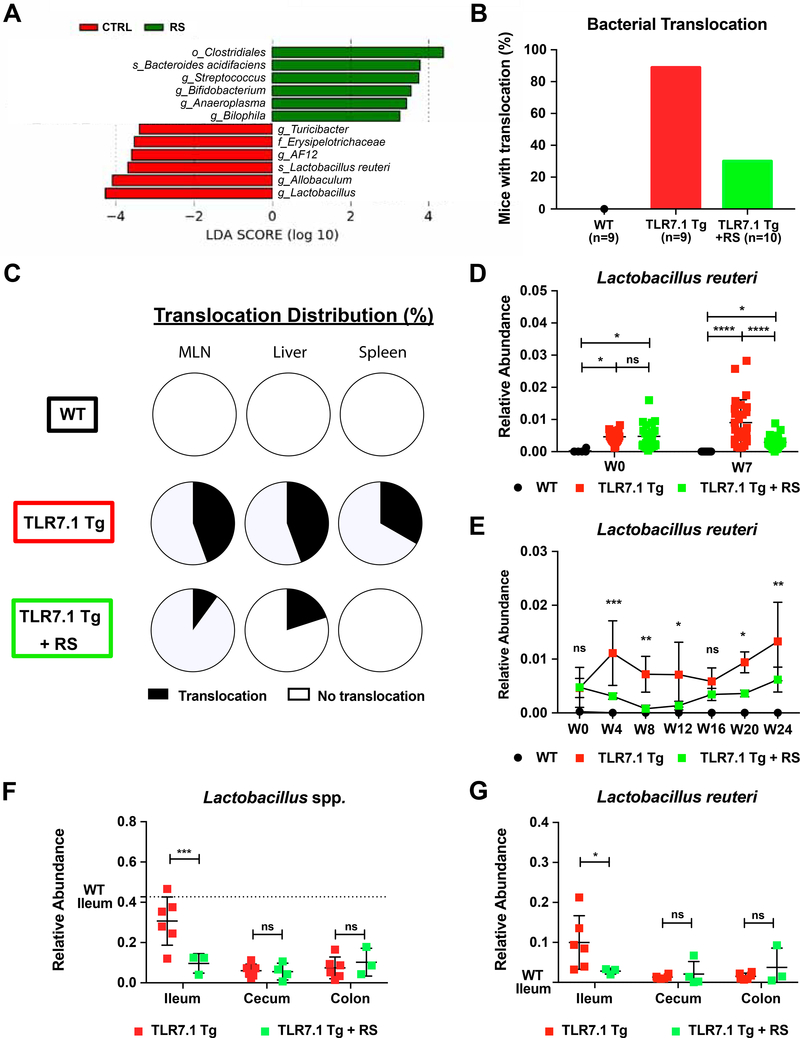
Figure 5. Resistant starch modulates the gut microbiome and decreases L. reuteri translocation.
Fecal pellets and gut segments from WT C57B1/6, TLR7.1 Tg, and TLR7.1 Tg mice fed with resistant starch (RS) were collected; bacterial DNA was isolated and sequenced. (A) LDA scores of altered taxa in TLR7.1 Tg mice fed with RS compared to control (CTRL) diet. Shown are significantly altered taxa (increased in green, decreased in red). (B) Total percentage of bacterial translocation as determined by culture. (C) Distribution of Lactobacillus spp. translocation in MLN, liver, and spleen from WT, TLR7.1 Tg, and TLR7.1 Tg mice fed with RS. Relative abundances of fecal L. reuteri (LR) in TLR7.1 Tg mice fed with RS compared to CTRL diet evaluated by 16S rDNA gene sequencing in (D) experimental or (E) survival cohorts. (F) Relative abundances of Lactobacillus spp. and (G) LR in gut tissues evaluated by 16S rDNA gene sequencing. The results are expressed as mean ± SEM (n=3–20 mice per group). Data are representative of at least three independent experiments. *P<0.05 was considered statistically significant; **P <0.01; ***P<0.001; ****P<0.0001; ns=not significant; W0=week RS begins. See also Figure S5 and Tables S4-S5.