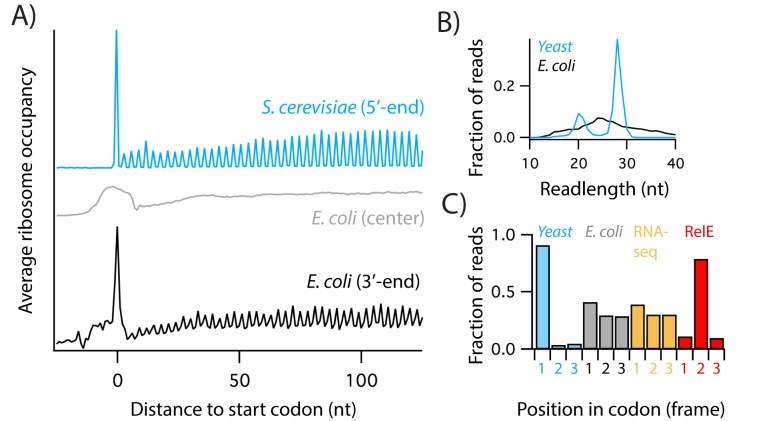
Cross-correlation plots were generated by first, calculating a genome-wide map of SD affinity using an eight nt sliding window, and then taking the correlation of the SD-affinity map with ribosome density at different offset values (
Li et al., 2012). The strong peak at −22 in L18 (black) indicates a positive correlation between SD affinity and ribosome density as would be expected if SD motifs caused ribosomes to pause in ORFs. The −22 offset is consistent with the known spacing between the SD motif and the 3’-boundary of the ribosome during initiation (top). In contrast to the strong positive correlation at −22 seen in L18, a negative correlation is observed in L17 (blue). This difference arises from how footprints were isolated: L18 contains exclusively long RPFs (>28 nt) whereas L17 contains exclusively short RPFs (20–30 nt). Given that footprints that interact with rRNA tend to be longer (30–35 nt), isolating only long RPFs leads to artificial enrichment of ribosome density at SD-like motifs. No SD pauses are observed in our libraries (e.g. L26, red) that capture the whole distribution of footprint sizes, nor were they observed in samples prepared using our new methods with high MgCl
2 lysis buffers with cells harvested by either filtration (L29) or direct freezing (L33) (purple and green respectively). In addition to the peaks at −22 initially attributed to pauses on SD motifs, two other peaks are also observed. The peak at −15 arises from pauses at Gly codons (
Figure 3 and
Mohammad et al., 2016) because Gly codons are G-rich, giving a spurious but strong SD affinity. In a similar fashion, the peak at 0 arises from cloning bias because the nucleotide G is enriched at the 3’-ends of reads.