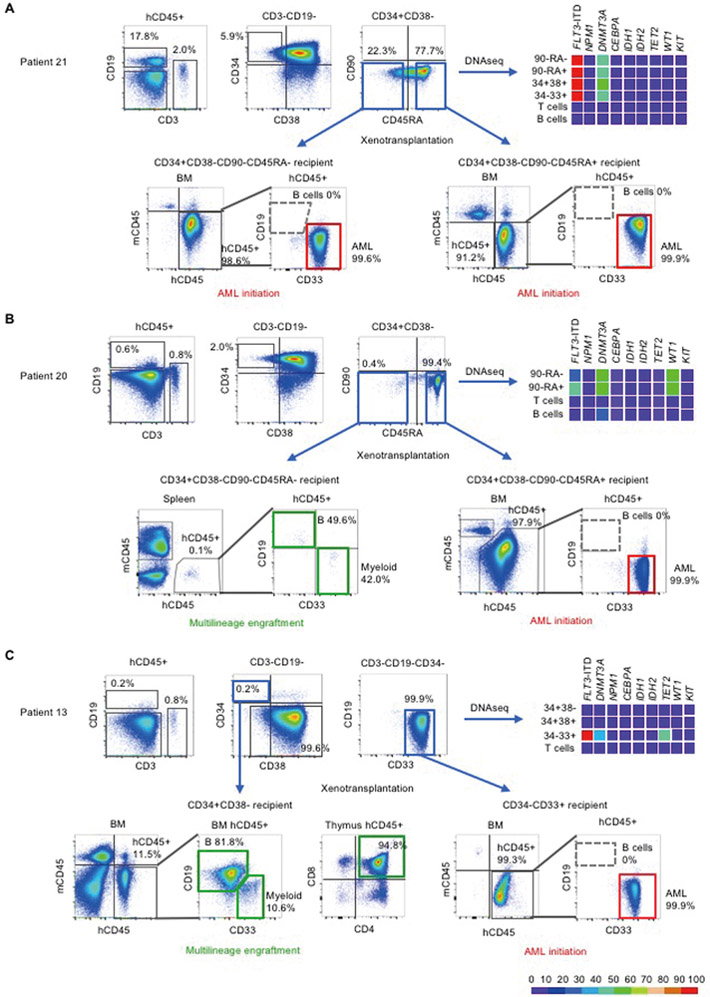
Figure 2.
Mutational profiles and in vivo engraftment of patient-derived subpopulations
Three additional patients are shown in fig. S1. In each patient sample, human CD45+CD3+CD19- T and human CD45+CD3-CD19+ B cells were identified. Within CD3-CD19- non-T non-B cells, subpopulations were identified based on CD34, CD38, CD90, and CD45RA surface expression. These populations underwent PCR for FLT3-ITD mutation and DNA sequencing for the other genes indicated. Variant allele frequencies are shown as heat maps. In Patient 21 (A) and Patient 20 (B), the CD34+CD38-CD90-CD45RA- and CD34+CD38-CD90-CD45RA+ subpopulations were identified. The in vivo fates of CD34+CD38-CD90-CD45RA- subpopulations differed between patients 20 and 21, showing engraftment with multilineage human hematopoiesis in Patient 20 (indicated by green rectangles) but initiation of AML in Patient 21 (indicated by red rectangles). In Patient 13 (C), the CD34+CD38- subpopulation showed multilineage repopulation, whereas CD34-CD33+ subpopulation with additional FLT3-ITD and NPM1 mutations initiated AML. AML engrafted recipients showed no B cell engraftment (indicated by gray dashed outlines on flow cytometry plots). Detailed information on variants found in each patient is shown in table S2.