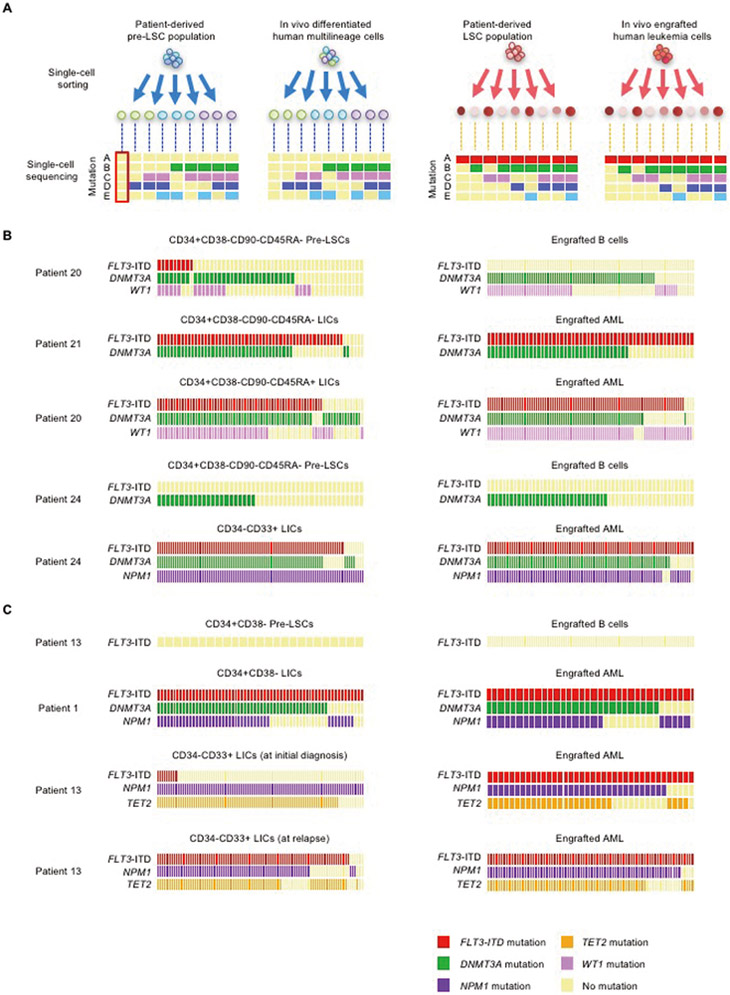
Figure 3.
Mutations contributing to distinct in vivo cell fates identified by single-cell functional genomics
(A) Patient-derived subpopulations with defined in vivo fates and their in vivo progeny underwent single-cell mutation profiling. Through this strategy, mutations present in patient-derived pre-leukemic stem cells and LIC clones were tracked and linked to in vivo fates. (B,C) Using samples from five patients, patient-derived multilineage-engrafting and LIC-containing population and engrafted B cells and AML cells were subjected to single-cell DNA sequencing for variants detected in each indicated gene in each patient. FLT3-ITD sequences with highly variable repeated sequence patterns were detected by single-cell PCR. In (B) and (C), each column of rectangles represents an individual cell. The presence or absence of mutations in each gene is shown by colors of rectangles as indicated.