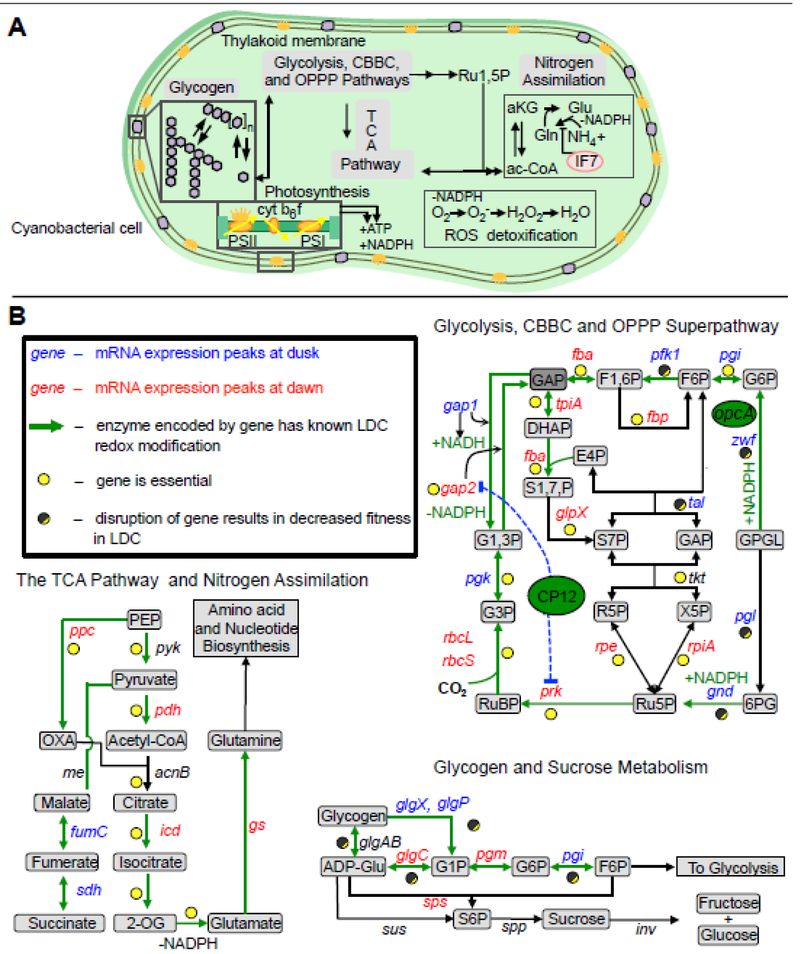
Figure 1. Shift work: snapshot of cellular activities across the day and night.
A) A representation of an S. elongatus cell with an overview of the major metabolic pathways important for day-night physiology. B) Reactions of glycogen metabolism, TCA pathway, and central metabolism that are present in S. elongatus are provided in more detail. Green arrows in panel B indicate reactions where the respective enzyme has a detectable light/dark-dependent redox modification in Synechocystis sp. PCC 6803 [69]. Data about the peak expression time of circadian genes is indicated using data collected from S. elongatus PCC 7942 [9]. Genes colored in red peak in expression in the subjective morning, genes colored in blue peak in expression in the subjective evening, and genes colored in black have no detectable circadian rhythm in S. elongatus. Essential genes and genes that cause light-dark sensitive phenotypes when mutated are indicated by full yellow and half yellow-half dark grey circles, respectively. Abbreviations: PBS, phycobilisome; PSII, photosystem II; cyt b6f, cytochrome b6f; PSI, photosystem I; Fd(red), ferredoxin (reduced); Ru1,5P, ribulose-1,5-bisphosphate; 3PG, 3-phosphoglycerate; 1,3-BPG, 1,3-bisphosphoglycerate; GAP, glyceraldehyde-3-phosphate; F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; G1P, glucose-1-phosphate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; ac-CoA, acetyl-CoA; aKG, a-ketoglutarate; Glu, glutamate; Gln, glutamine.