Significance
Plants, like animals, have complex disease resistance systems in which receptors interact directly or indirectly with effectors of disease produced by pests and pathogens. To minimize the fitness cost of these systems to the plant, there are miRNAs that target the mRNAs of a family of receptor proteins required for disease resistance. Target site mimics of these miRNAs confer enhanced quantitative resistance in tomato against an oomycete and a bacterium. These findings are consistent with a role of the receptor proteins in quantitative disease resistance and show how blocking these miRNAs could be a useful approach in crop protection.
Keywords: microRNA, NLR, siRNA, quantitative disease resistance, noncoding RNA
Abstract
Nucleotide binding site leucine-rich repeat (NLR) proteins of the plant innate immune system are negatively regulated by the miR482/2118 family miRNAs that are in a distinct 22-nt class of miRNAs with a double mode of action. First, they cleave the target RNA, as with the canonical 21-nt miRNAs, and second, they trigger secondary siRNA production using the target RNA as a template. Here, we address the extent to which the miR482/2118 family affects expression of NLR mRNAs and disease resistance. We show that structural differences of miR482/2118 family members in tomato (Solanum lycopersicum) are functionally significant. The predicted target of the miR482 subfamily is a conserved motif in multiple NLR mRNAs, whereas for miR2118b, it is a noncoding RNA target formed by rearrangement of several different NLR genes. From RNA sequencing and degradome data in lines expressing short tandem target mimic (STTM) RNAs of miR482/2118, we confirm the different targets of these miRNAs. The effect on NLR mRNA accumulation is slight, but nevertheless, the tomato STTM lines display enhanced resistance to infection with the oomycete and bacterial pathogens. These data implicate an RNA cascade of miRNAs and secondary siRNAs in the regulation of NLR RNAs and show that the encoded NLR proteins have a role in quantitative disease resistance in addition to dominant gene resistance that has been well characterized elsewhere. We also illustrate the use of STTM RNA in a biotechnological approach for enhancing quantitative disease resistance in highly bred cultivars.
Nucleotide binding site leucine-rich repeat (NLR) proteins of plants are central components of the innate immune system that protects against pests and pathogens. These proteins are encoded in multigene families and they regulate signal transduction pathways leading to disease resistance (1). There are several negative regulators of these resistance pathways that, presumably, reduce the likelihood that disease resistance is activated in the absence of a pest or pathogen. Such pathogen-independent induction could be damaging to the host because the disease resistance mechanisms may be associated with programed cell death called hypersensitive response, changes to the cell wall, production of active oxygen species, and strong activation of pathogenesis-related genes that compromise reproductive fitness. Transgenic lines overexpressing NLRs (2–4) and gain-of-function mutations, like suppressor of npr1-1, constitutive 1 (5) or suppressor of salicylic acid insensitive 4 (6), illustrate how inappropriate activation of disease resistance can damage the plant. In normal conditions the negative regulators may mitigate the potential cost of NLR gene diversification and they could be particularly beneficial to plants with many NLR genes (7).
The importance of these negative regulators is reflected in their diversity. There are, for example, suppressor proteins affecting either the NLR proteins themselves or components of the downstream signal transduction pathways (8, 9). There are also miRNAs and siRNAs that function as negative regulators of NLR mRNA (10–13). These small RNAs (sRNAs) bind to their target mRNA by Watson–Crick base pairing, and they silence the expression of the mRNA-encoded protein through various mechanisms affecting RNA stability or translation in which the effectors are proteins of the Argonaute family (14).
Among the miRNA regulators of NLRs (7), the miR482/2118 family is the most diverse. This family is present in modern seed plants, although, in some instances, the apparent conservation may reflect repeated rounds of convergent evolution (15). The miR482/2118 family members are all 22 nt rather than the more usual 21 nt in length (10–12), and the additional nucleotide is functionally significant because it influences the fate of the targeted mRNA: targets of 21-nt miRNA are simply degraded by the Argonaute nuclease activity (14), whereas 22-nt miRNA targets are converted into a dsRNA by the RNA-dependent RNA polymerase 6. The dsRNA is then cleaved by a DCL protein to generate an array of 21-nt secondary siRNAs that may be phased with respect to the binding site of the miRNA (16, 17). As a result of this process there is the potential for 22-nt miRNAs to establish regulatory cascades in which mRNAs are targeted by both primary miRNAs and secondary sRNAs.
Here we focus on the miR482/2118 family in tomato and we set out to establish the extent to which they influence NLR RNAs and disease resistance. Our approach involved the use of short tandem target mimic (STTM) RNAs that would inactivate the miR482/2118 family members and prevent primary or secondary regulation of NLR mRNAs. Our findings confirm that, although the miR482 and miR2118 subfamilies have similar sequences, they are functionally distinct in tomato. The miR482 subfamily targets NLR mRNAs as already described (10–12) and triggers secondary siRNA production. It is possible, therefore, that NLR RNAs could be both primary and secondary targets of the miR482 family. One isoform of miR2118—miR2118b—is likely to influence NLRs through secondary siRNAs. Its main primary target is not an NLR but a long noncoding RNA—TAS5—from which secondary sRNAs are produced that could target NLR coding sequence RNAs. This primary and secondary negative regulation of NLR RNAs by miR482/2118b has an effect on quantitative disease resistance because the STTM lines are less susceptible to an oomycete and a bacterial pathogen than control lines. This enhanced resistance was achieved without a large effect on growth and development of the plants, and it may be useful to protect tomato and other species against pests and diseases.
Results
Revisiting the miR482/2118 Family and Their Targets in Tomato.
There are numerous families of miRNAs that negatively regulate defense by targeting conserved motifs in NLR RNAs. The miR482/2118 family is the most extensive of these families and it is present in most lineages of seed plants due to either conservation or convergent evolution (7). This analysis focused on the five members of this family (SI Appendix, Table S1) that (i) align to the tomato genome and (ii) feature in our sRNA datasets from leaves of 1-mo-old tomato plants.
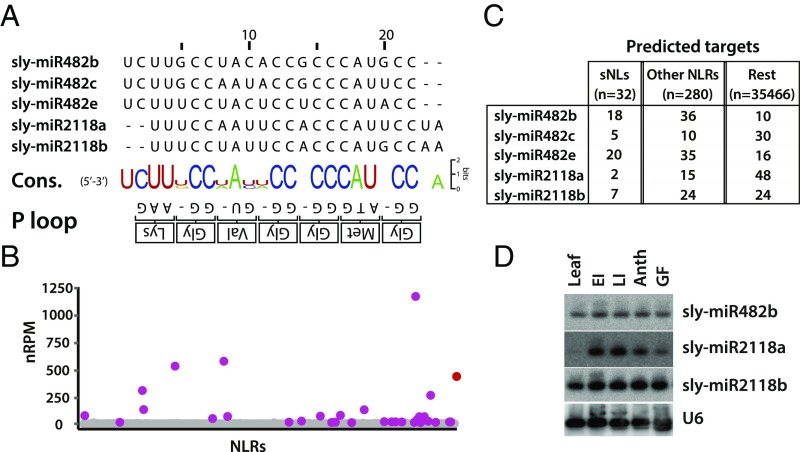
The sequences of these five miRNAs (Fig. 1A) are complementary to the RNA representation of the conserved P-loop motif (GMGGVGKT) in NLR proteins with sequence variation at 5/6 variable sites corresponding to wobble positions. This pattern indicates that this small family of miRNAs could target a larger number of NLR RNAs with synonymous coding sequence variation (12). Using refined prediction algorithms (18) we estimate that individual miR482/2118 species could potentially target between 15 and 55 NLR RNAs (Fig. 1B and SI Appendix, Table S2).
Fig. 1.
The miR482/2118 family in tomato. (A) Nucleotide sequence alignment of mature miR482/2118 members in tomato. The consensus sequence of each position in the alignment vs. the P-loop motif is shown at the bottom. (B) Dot plot representing the sum of 21-nt sRNA nRPM aligning to an individual NLR. A total of 32 NLRs presented >10 nRPM counts and were defined as sNLs (purple). TAS5 (red) is added as a reference. (C) Summary of target prediction of all miR482/2118 members. (D) RNA gel blot analysis of tomato miR482/2118 members in various tissues of plant development. Column 4 shows the same blot hybridized with U6 as a loading control. Anth, anthesis; EI, early influorescence; GF, green fruit; LI, late influorescence.
The miR482/2118 family members also have the potential to trigger secondary siRNA production on their mRNA targets (16) because they are 22 nt in length (10–12); correspondingly, there were 32 NLR genomic regions with 10 or more 21- to 22-nt siRNA normalized reads per million (nRPM) mapping (Fig. 1C) from our sRNA datasets. Other studies in different species refer to these NLRs with overlapping siRNAs as phasi-NLRs (10, 15, 19), although the extent of phasing may be low. We use the term sNL here referring to NLR mRNAs with secondary sRNAs that may or may not be phased. Most of these tomato sNLs in our datasets are predicted targets of miR482s with 30 of 32 of them encoding nucleotide binding site leucine-rich repeats with coiled coil domains (CNLs) rather than nucleotide binding site leucine-rich repeat with Toll/interleukin-1 receptor homology domains (TNLs) at the amino terminus (Fig. 1B and SI Appendix, Table S3).
Compared with miR482 predictions, potential targets of miR2118s were less enriched for NLRs (Fig. 1B and SI Appendix, Table S2) or secondary siRNAs. One of the two isoforms miR2118a—like its close homologue in other species (20, 21)—was much less abundant in leaf tissues than in flower and fruit (Fig. 1D) and it may not have a large effect on NLR mRNAs. In contrast, miR2118b, like miR482 isoforms, was abundant in leaves and reproductive tissues (Fig. 1D) (12) and it can potentially target several RNAs, including seven sNLs (Fig. 1B and SI Appendix, Table S2) and an RNA species that had been identified previously as TAS5 (22).
Our analysis reveals that TAS5 is atypical of NLR RNAs, because it has sequence similarity to both CNL and TNL types of NLR RNA on both the sense and antisense orientations (SI Appendix, Fig. S1). It is unlikely to be translated into a functional protein [calculated coding potential score of −1.146 (23, 24)], and a more likely interpretation is that TAS5 is a long noncoding RNA composed of rearranged and degenerated sequences from multiple NLR genes (SI Appendix, Fig. S1). It has two miR2118b target sites that were detected bioinformatically and by degradome analysis (SI Appendix, Fig. S2 and Tables S2 and S4), and the siRNAs are predominantly in a phased register. This phasing pattern starts at or adjacent to the more 5′ miR2118b target sites (SI Appendix, Fig. S2).
These findings indicate that there are both structural and functional differences between the miR482 and miR2118 subfamilies. The miR482 subfamily (12) targets the CNL NLRs directly and may also have an indirect effect on NLR RNAs via secondary siRNAs. The miR2118 subfamily, in contrast, is different in that its alignment with the P-loop coding motif is shifted by two nucleotides and is only associated with secondary siRNAs (Fig. 1C) in a few instances. The miR2118a variant is likely to have little if any effect on NLRs. The miR2118b, in contrast, may act via TAS5 secondary siRNAs on both TNL- and CNL-type NLRs.
Target Mimics Block Production of Secondary NLR siRNAs.
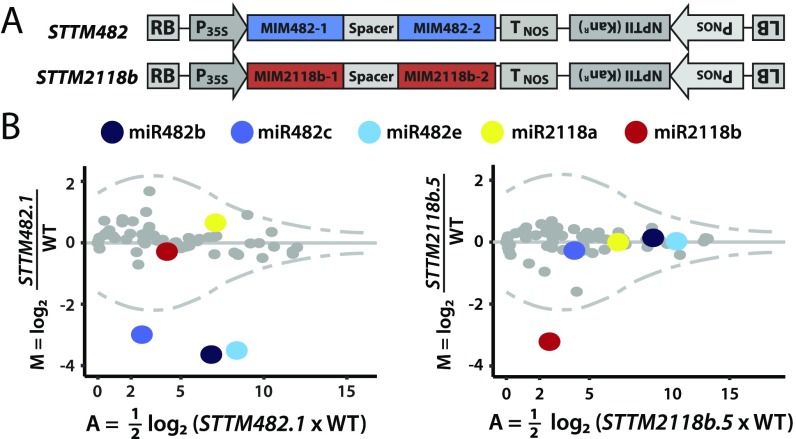
To further investigate the function of miRNAs from the miR482/2118 family in tomato, we generated STTM transgenes. They were expressed from the strong 35S promoter and were designed so that their transcripts would inactivate the miRNAs 482 or 2118b. We did not test miR2118a, as it is expressed predominantly in reproductive tissue (Fig. 1D). The design of the STTM constructs was based on the IPS1 natural target mimic of miR399 (25) with two tandem target sites (Fig. 2A) to enhance efficiency (26). These target mimic sequences had a 3-nt bulge in the miRNA/target RNA duplex structure that would prevent cleavage of the target mimic RNA and channel the miRNAs away from their natural targets.
Fig. 2.
STTMs inactivate specific miRNAs and inhibit production of secondary siRNAs. (A) Diagram of target mimic constructs used in this study. (B) MA plot showing fold changes of miRNAs in STTM lines. Tomato mature miRNA sequences were extracted from miRBASE. The blue dots indicate miR482, yellow is miR2118a, and red is miR2118b; gray indicates other miRNAs. The dotted lines represent a Poisson distribution with 1% significance values at the top and bottom of the range, applying the 0 correction (if nreads = 0; +1). sRNA reads are normalized to the whole library with nRPMs and presented as the mean from three biological replicates.
Lines transformed with the STTM482 and STTM2118b constructs were screened for transgene expression and effects on target miRNAs. Of 16 stable lines (8 per construct), one-half showed reduced levels of the corresponding miRNAs (SI Appendix, Fig. S3). From these lines, for subsequent detailed analysis, the top inactivating lines for STTM482 (lines 1 and 3 in SI Appendix, Fig. S3) and STTM2118b (lines 5 and 7 in SI Appendix, Fig. S3) were carried to the further generations. In 1-mo-old T2 plants, high-throughput sequence analysis of miRNAs in STTM482.1 revealed a specific 10-fold reduction over the WT in all miR482 subfamily members, whereas in STTM2118b.5, the effect was exclusively on miR2118b (Fig. 2B and SI Appendix, Fig. S4). All other miRNAs were similarly abundant in the STTM lines and WT plants. The effect of the STTMs, therefore, was specific for the cognate miRNA.
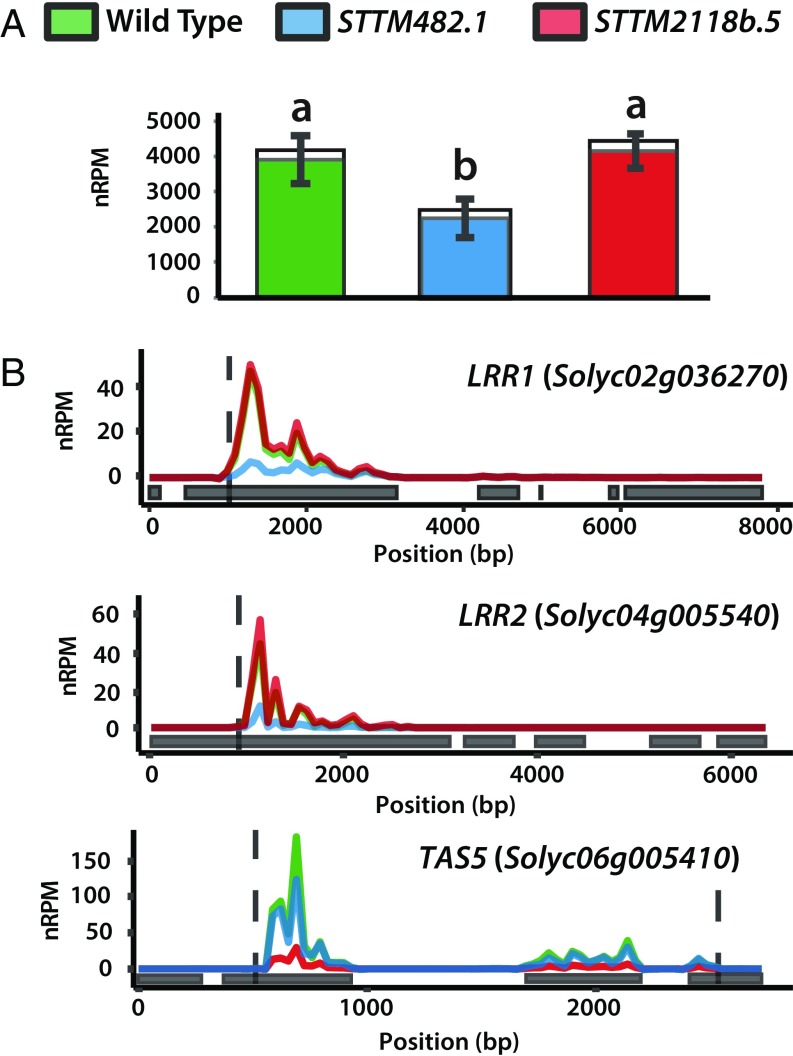
There was an additional effect of the STTMs on sRNAs other than miRNAs. To detect this effect, we aligned sRNA reads to the tomato genome and identified differential small RNA loci (DSLs) using the segmentSeq and baySeq packages (27). DSLs in STTM482.1 were enriched for sNLs, and most of them (10 of 14) overlapped with predicted targets of miR482 members, whereas in STTM2118b.5, the only DSL was TAS5 (SI Appendix, Table S6). The siRNA levels at 14 DSL sNLs were at least twofold higher in the WT than in STTM482.1 (SI Appendix, Table S7). At LRR1 (Solyc02g036270) and LRR2 (Solyc04g005540), two well-studied CNL RNAs targeted by miR482 (12), there were abundant siRNAs aligned to the 3′ side of the miRNA cleavage sites (Fig. 3B) that were fourfold reduced in the STTM482.1 but not in STTM2118b.5. Conversely, there was a fivefold reduction in TAS5-derived siRNAs in STTM2118b.5 but not in STTM482.1 (Fig. 3B and SI Appendix, Table S7).
Fig. 3.
miR482- and miR2118b-mediated silencing is relieved in STTM482 and STTM2118b lines, respectively. (A) Bar plot of total 21-nt siRNA counts in NLRs in WT and STTM lines. Colored fractions correspond to sNLs. Statistically significant differences were found using the one-way ANOVA test followed by Tukey’s honestly significant difference (HSD) test at 95% confidence limits. (B) Abundance of 21-nt siRNAs along two sNLs (LRR1 and LRR2, two well-studied CNL genes targeted by miR482) and TAS5 loci. Positions corresponding to the cleavage sites of their miRNA triggers (miR482s and miR2118b, respectively) are indicated by dashed lines. Gray boxes indicate the position of exons. sRNA reads are normalized to the whole library with nRPMs and presented as the mean from three biological replicates.
Target Mimic Effects on NLR mRNA Accumulation.
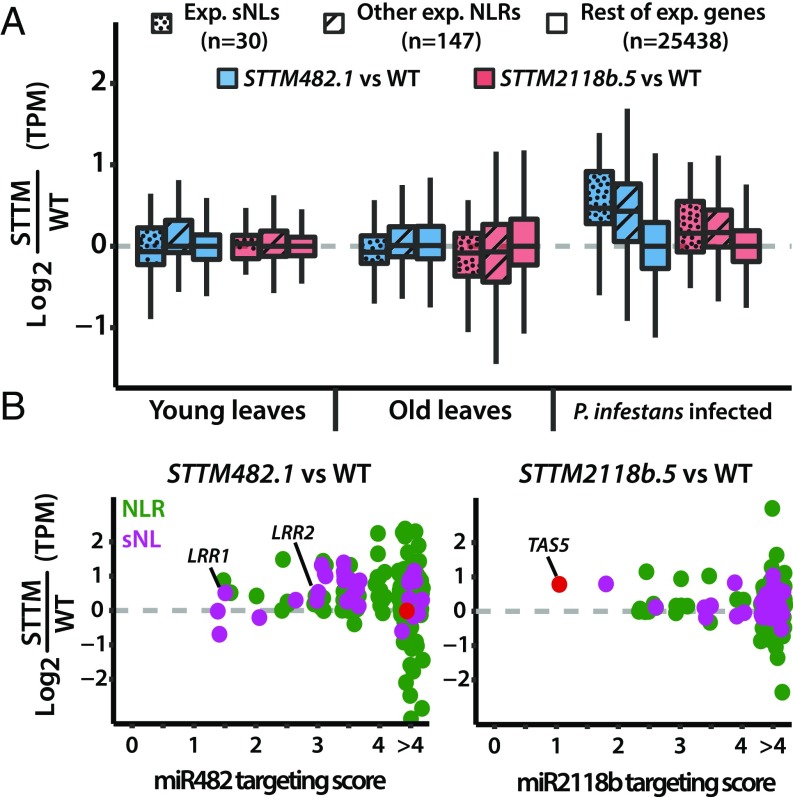
We further investigated the effect of miR482/2118 species on the NLR mRNAs using RNAseq data of WT and T2 STTM lines. We used young and old leaves and detached leaves inoculated with zoospore droplets of Phytophthora infestans 88069. The analysis of these datasets revealed that the overall effects of the STTM RNAs were small in all of the conditions tested. The expressed sNLs were more affected than other NLRs or other genes, but only in the infected plants was the trend toward elevated expression (Fig. 4A). The previously described miR482 targets LRR1 and LRR2 and a few other sNLs and NLR RNAs showed consistent small up-regulation (1.5- to 2.5-fold changes) in all conditions in STTM482 but not STTM2118b lines, and TAS5 was consistently up-regulated in STTM2118b (SI Appendix, Table S7) but not in the STTM482 lines. These observations were validated using qRT-PCR (SI Appendix, Fig. S5).
Fig. 4.
Overall NLR expression is increased in STTM482 and STTM2118b lines during biotic stress. (A) Box plot of transcript abundances differences between STTM lines and the WT across different conditions. Colors correspond to STTM482.1 (blue) and STTM2118b.5 (red) vs. the WT. Dotted patterns represent expressed sNLs, whereas other NLRs are represented as striped boxes, and plain boxes represent the rest of the genes in the tomato genome. (B) Scatter plot representing changes in abundance against the targeting score by either miR482 or miR2118b for each of the expressed sNL (violet) or other NLR (green) in STTM lines vs. the WT during P. infestans infection. TAS5 (red) was added as a reference. In all instances, RNA abundances are calculated as transcripts per million (TPM) and presented as the mean of six biological replicates.
To find out whether the small up-regulation effect was due directly to reduced targeting by the respective miRNAs, we focused on the P. infestans-inoculated samples and we compared the targeting score of either miR482 or miR2118b with the degree of overexpression for each of the sNLs and expressed NLRs (SI Appendix, Table S2). There was no correlation (Fig. 4B); we conclude from these data (Figs. 3 and 4) that the miR482/2118 species are regulators of NLR expression, but to account for the lack of a simple correlation of targeting score and mRNA accumulation, there may be additional mechanisms involved as discussed below.
Inactivation of miR482 and miR2118b Enhances Resistance to P. infestans and Pseudomonas syringae.
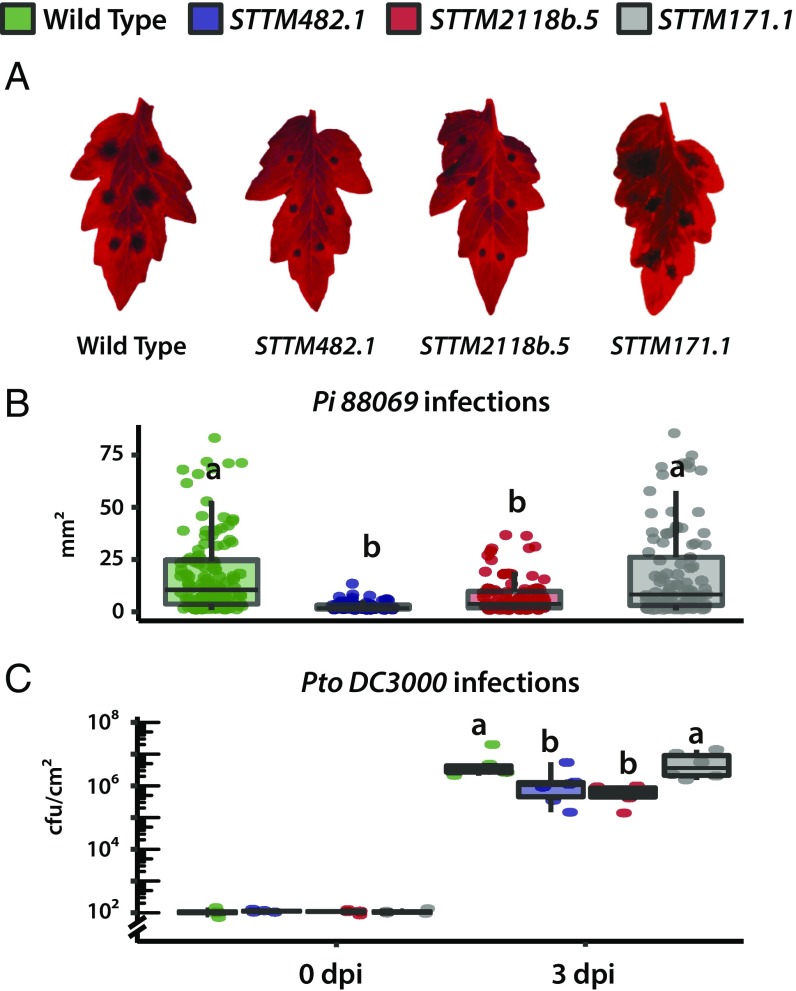
To investigate the effect of miR482/2118 on disease resistance, we inoculated detached leaves of T2 STTM lines and WT plants with zoospore droplets of the P. infestans 88069. The tomato cultivar M82 (LA3475), from which all of the plants in this study are derived, is highly susceptible to P. infestans (28). We measured the size of necrotic lesions to measure progression of the disease, and according to this criterion, the STTM482.1 and STTM2118b.5 were less susceptible than the nontransgenic control plants with significantly smaller lesion size at 3 d postinfection (dpi) (Fig. 5 and SI Appendix, Fig. S4). As a transgene control, a transgenic line expressing an STTM against miR171 (line STTM171.1), an miRNA with no reported role in plant defense, was used and was as susceptible as nontransgenic lines.
Fig. 5.
Sequestration of miR482 and miR2118b influences broad spectrum resistance to pathogens. (A) Representative images of detached leaves under blue light 3 dpi with P. infestans 88069. (B) Box plot and leaf images of lesion size in WT and mimicry lines. Statistically significant differences were determined using the one-way ANOVA test followed by Tukey’s honestly significant difference (HSD) test at 95% confidence limits (n = 12). (C) Box plot of bacterial population in WT and STTM line leaves infected with Ps. syringae pv. tomato DC3000. Bacterial counts at 0 and 3 days postleaf infiltration. Statistically significant differences were determined using the ANOVA test followed by Tukey’s HSD test at 95% confidence limits (n = 6). Both infection experiments were repeated three times with similar results.
We also tested the effect of the STTM constructs on resistance against Ps. syringae pv. tomato DC3000. Bacterial titers in inoculated leaves attached to the plant were lower at 3 dpi in both STTM482.1 and STTM2118b.5 than WT and STTM171.1 lines (Fig. 5C and SI Appendix, Fig. S4). This effect indicates that the primary or secondary targets of miR482s and miR2118b have a role in antibacterial immunity.
Defense activation may be at the expense of the plant’s fitness (29). In the STTM lines, however, general growth was similar in the control and STTM lines used in the disease resistance tests other than a small decrease in shoot length in STTM482.1 (SI Appendix, Fig. S6). These tests may not be sensitive to other slight differences in fitness but they indicate that enhanced disease resistance in the STTM lines was not associated with gross effects on growth and development.
Discussion
miR482/2118b as Regulators of NLRs and Quantitative Disease Resistance.
In this paper, we describe definitive evidence that miR482/2118b family members are negative regulators of quantitative disease resistance in tomato: suppression of miR482 and miR2118b using STTMs enhanced resistance against both P. infestans and Ps. syringae (Fig. 5). These findings are consistent with hypersusceptibility to Verticillium and P. infestans caused by overexpression of miR482 family members (30, 31). Similarly, higher expression levels of miR482/2118 members correlated with hypersusceptibility to P. infestans (32) in a separate study, and their expression level correlated inversely with resistance against Fusarium oxysporium (33). Our findings also confirm the very recent report that STTM482b confers enhanced resistance against P. infestans (31).
To explain the quantitative disease resistance in the STTM lines (Fig. 5) there could be, in principle, mechanisms that either involve NLRs as miR482/2118b targets or are NLR independent. We do not favor the NLR-independent mechanism, however, because known defense-related non-NLR mRNAs do not undergo large increases in abundance in the STTM lines (SI Appendix, Fig. S8 and Table S8) and there are very few predicted miR482/2118b targets that are not NLR RNAs (SI Appendix, Table S2). One exception is a predicted target of miR2118b encoding a DnaJ-type chaperone protein (Solyc01g105340) (SI Appendix, Table S2), but this RNA is not differentially expressed in the STTM lines (SI Appendix, Table S8) and its encoded protein is not easily accommodated in models of quantitative disease resistance. For these reasons, we favor quantitative disease resistance in the STTM lines that is mediated by NLRs, although we are aware that additional tests are required to block NLR signaling pathways or to knock out parts of the TAS5 locus and sNL loci.
How many NLRs might affect the quantitative resistance in the STTM plants? With miR482, there are at least 32 NLRs generating secondary siRNAs (Fig. 1) and, in principle, there could be many more implicated in a miR482/2118b cascade. The first layer in this cascade would involve NLR RNAs that are direct targets of miR482 and a second layer might involve NLR targets of secondary siRNAs. With miR2118b, however, the main primary target generating secondary siRNA is a noncoding RNA—TAS5. It is unlikely that this TAS5 RNA affects resistance directly, and it is likely that NLRs are targeted by the secondary siRNAs that are reduced in the STTM2118b.5 (Fig. 3 and SI Appendix, Fig. S2 and Tables S4–S7).
Well-described RNA silencing mechanisms involving siRNA and miRNA result in cleavage and degradation of the target RNAs (14), and we had expected that the sNL RNAs and other NLR RNA targets of miR482/2118b family would increase in abundance in the STTM lines. There was, however, only a small increase (Fig. 4 and SI Appendix, Fig. S6 and Table S7) even for the sNL RNAs with reduced secondary sRNAs in the STTM lines (Fig. 3). To reconcile these RNAseq results (Fig. 4 and SI Appendix, Fig. S6 and Table S7) with NLR-mediated quantitative resistance (Fig. 5), we hypothesize that there is a translational effect in the STTM plants that is not associated with major changes in the abundance of the targeted NLR RNAs. An effect in tomato of miR482 family members on NLR translation is also supported by an independent report (33).
NLR genes are normally associated with dominant gene resistance and with mechanisms that confine the pathogen to the site of initial infection. This dominant gene resistance was the first described phenotype of NLRs (34), and it is normally specific for races or strains of the pathogen. The NLRs in this race-specific resistance mediate direct or indirect recognition of effectors that are transported into the infected cell (1). This “effector-triggered immunity” then triggers metabolic and molecular changes that hinder the progression of disease (35). Our tests were not, however, with race-specific resistance. The plant genotypes were fully susceptible to both pathogens, and the STTM-mediated effect was manifested as quantitative resistance rather than complete suppression of the pathogen (Fig. 5).
To explain this quantitative resistance in the STTM plants, we propose that there is a low level of recognition of pathogen effectors, even in susceptible plants, but that any triggering of resistance would be too slow or too weak to prevent accumulation and spread of P. infestans and Ps. syringae. In the STTM plants, however, the reduced silencing of NLRs may allow stronger recognition and more rapid activation of defense. Alternatively, any increased expression of NLRs in the STTM lines could cause autoactivation of disease resistance in the absence of elicitor recognition, analogous to the effects of overexpressing NLRs in noninfected plants (2–4).
Evolutionary Dynamics of NLR Regulation by miRNA.
The miRNA regulation of NLRs is highly variable in angiosperm species. In addition to the miR482/2118 family, there are numerous other miRNAs, including miR5300, miR6024, miR6026, miR825, and others, with the potential to target NLRs (36). All of these miRNAs are variably present depending on the species, indicating a dynamic evolutionary process in which RNA silencing of NLR RNAs is lost and gained (7, 37).
The miR2118 family is a special case of NLR regulation that is dependent on two genome events. One of these changes in the Solanaceae involves functional diversification of miR2118 members, with one of them (miR2118b) being expressed in the vegetative phase including leaves and another (miR2118a) having the pattern that is typical of most monocot and dicot species (Fig. 1) in reproductive phases (20, 21). Consistent with a previous report (38) we were unable to find homologues of miR2118b outside of the Solanum clade (SI Appendix, Fig. S7). A second Solanaceae-specific genome change (SI Appendix, Figs. S1 and S7) would have been a rearrangement of TNL and CNL NLR genes, resulting in the TAS5 locus. These changes illustrate how the evolutionary dynamics of NLR regulation are not only dependent on loss or gain of the miRNA genes. There can also be neofunctionalization of miRNA genes, as with miR2118b, and the addition of noncoding RNAs into the RNA silencing cascade, as with TAS5.
We have speculated previously that the miR482-mediated down-regulation of NLRs is a process that allows the plant to trade off the costs and benefits of NLRs (37). Accumulation of NLR proteins has a cost that is presumably related to metabolic changes associated with disease resistance and a benefit due to protection against pests and pathogens. The balance of costs and benefits may depend on the ecological niche occupied by the plant and on the effectiveness of other defense systems.
A component of our earlier speculation was that pathogen-derived suppressors of RNA silencing would relieve the miR482-mediated suppression of NLRs in infected plants (12). We originally envisioned that this process would operate in virus-infected plants (14) but with the identification of such suppressors encoded by other pathogens, including P. infestans (39), it could be more general. The effect of a suppressor of silencing could explain the increase of NLRs in P. infestans-infected plants (Fig. 4A).
To explain the apparent minimal cost of STTMs on growth and development of the plants, we suggest that the protected glasshouse environment of our tomato plants allowed them to tolerate the increased expression of NLRs without effects on growth and development of the plant (SI Appendix, Fig. S6). There may also be other changes associated with the domestication and breeding of cultivated tomato that protect the plant against the costs of increased NLRs. Additional testing will be required to establish whether there is a cryptic growth phenotype associated with STTM expression and also, whether the basal immunity can be enhanced further. It could be that a combination of the two STTMs would have a stronger effect than the individual species. Alternatively, the enhanced basal immunity would be useful if it is part of an integrated management strategy to protect crops against disease.
Materials and Methods
Plant strains, plasmid constructs, Northern blots, bioinformatic analyses, and pathogen tests are described in SI Appendix. The tomato (Solanum lycopersicum) cultivar M82 was used for all experiments. STTM transgenic lines were obtain via Agrobacterium tumefaciens-mediated stable transformation. Image analysis was performed on ImageJ. Statistical and bioinformatics analyses were performed using R.
Supplementary Material
Acknowledgments
We thank James Barlow, Mel Steer, Antonia Yarur, and Pawel Baster for technical and horticultural assistance; Stuart Fawke for maintaining and providing P. infestans 88069; Zhengming Wang for the STTM171.1 seeds; and Thomas Hardcastle for bioinformatics advice. This work was supported by the Balzan Foundation and European Research Council Advanced Investigator Grant ERC-2013-AdG 340642. S.S. is funded by Gatsby Foundation Fellowship GAT3395/GLD and Royal Society University Research Fellowship UF160413. D.C.B. is the Royal Society Edward Penley Abraham Research Professor. The funding bodies had no roles in the design of the study; the collection, analysis, or interpretation of data; or the writing of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the NCBI BioProject (accession no. PRJNA505207).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814380116/-/DCSupplemental.
References
- 1.Jones JD, Dangl J. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Oldroyd GED, Staskawicz BJ. Genetically engineered broad-spectrum disease resistance in tomato. Proc Natl Acad Sci USA. 1998;95:10300–10305. doi: 10.1073/pnas.95.17.10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F. Mutational analysis of the Arabidopsis nucleotide binding site–leucine-rich repeat resistance gene RPS2. Plant Cell. 2000;12:2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Dorey S, Swiderski M, Jones JDG. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 2004;40:213–224. doi: 10.1111/j.1365-313X.2004.02201.x. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Clarke JD, Zhang Y, Dong X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- 6.Shirano Y, Kachroo P, Shah J, Klessig DF. A gain-of-function mutation in an Arabidopsis Toll Interleukin1 Receptor–Nucleotide Binding Site–Leucine-Rich Repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell. 2002;14:3149–3162. doi: 10.1105/tpc.005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Xia R, Kuang H, Meyers BC. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol Biol Evol. 2016;33:2692–2705. doi: 10.1093/molbev/msw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, et al. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F, et al. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shivaprasad PV, et al. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halter T, Navarro L. Multilayer and interconnected post-transcriptional and co-transcriptional control of plant NLRs. Curr Opin Plant Biol. 2015;26:127–134. doi: 10.1016/j.pbi.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Galun E. RNA silencing in plants. In Vitro Cell Dev Biol Plant. 2005;41:113–123. [Google Scholar]
- 15.Xia R, Xu J, Arikit S, Meyers BC. Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol Biol Evol. 2015;32:2905–2918. doi: 10.1093/molbev/msv164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q, Han Z, Jiang H, Tian D, Yang S. Strong positive selection drives rapid diversification of R-Genes in Arabidopsis relatives. J Mol Evol. 2010;70:137–148. doi: 10.1007/s00239-009-9316-4. [DOI] [PubMed] [Google Scholar]
- 17.Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava PK, Moturu TR, Pandey P, Baldwin IT, Pandey SP. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics. 2014;15:348. doi: 10.1186/1471-2164-15-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arikit S, et al. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell. 2014;26:4584–4601. doi: 10.1105/tpc.114.131847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson C, et al. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19:1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhai J, et al. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci USA. 2015;112:3146–3151. doi: 10.1073/pnas.1418918112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Orban R, Baker B. SoMART: A web server for plant miRNA, tasiRNA and target gene analysis. Plant J. 2012;70:891–901. doi: 10.1111/j.1365-313X.2012.04922.x. [DOI] [PubMed] [Google Scholar]
- 23.Kong L, et al. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi X, Zhang Z, Ling Y, Xu W, Su Z. PNRD: A plant non-coding RNA database. Nucleic Acids Res. 2014;43:D982–D989. doi: 10.1093/nar/gku1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 26.Yan J, et al. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell. 2012;24:415–427. doi: 10.1105/tpc.111.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardcastle TJ, Kelly KA, Baulcombe DC. Identifying small interfering RNA loci from high-throughput sequencing data. Bioinformatics. 2012;28:457–463. doi: 10.1093/bioinformatics/btr687. [DOI] [PubMed] [Google Scholar]
- 28.Akhtar KP, et al. Evaluation of tomato genotypes for late blight resistance using low tunnel assay. J Plant Pathol. 2016;98:421–428. [Google Scholar]
- 29.Karasov TL, Chae E, Herman JJ, Bergelson J. Mechanisms to mitigate the trade-off between growth and defense. Plant Cell. 2017;29:666–680. doi: 10.1105/tpc.16.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, et al. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J Integr Plant Biol. 2015;57:1078–1088. doi: 10.1111/jipb.12348. [DOI] [PubMed] [Google Scholar]
- 31.Jiang N, Meng J, Cui J, Sun G, Luan Y. Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. Hortic Res. 2018;5:9. doi: 10.1038/s41438-018-0017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Vries S, et al. Expression profiling across wild and cultivated tomatoes supports the relevance of early mir482/2118 suppression for phytophthora resistance. Proc Biol Sci. 2018;285:20172560. doi: 10.1098/rspb.2017.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouyang S, et al. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014;10:e1004464. doi: 10.1371/journal.ppat.1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flor H. The complementary genic systems in flax and flax rust. Adv Genet. 1956;8:29–54. [Google Scholar]
- 35.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 36.Fei Q, Zhang Y, Xia R, Meyers BC. Small RNAs add zing to the zig-zag-zig model of plant defenses. Mol Plant Microbe Interact. 2016;29:165–169. doi: 10.1094/MPMI-09-15-0212-FI. [DOI] [PubMed] [Google Scholar]
- 37.González VM, Müller S, Baulcombe D, Puigdomènech P. Evolution of NBS-LRR gene copies among dicot plants and its regulation by members of the mir482/2118 superfamily of miRNAs. Mol Plant. 2015;8:329–331. doi: 10.1016/j.molp.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 38.De Vries S, Kloesges T, Rose LE. Evolutionarily dynamic, but robust, targeting of resistance genes by the miR482/2118 gene family in the solanaceae. Genome Biol Evol. 2015;7:3307–3321. doi: 10.1093/gbe/evv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong Q, et al. Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana. Mol Plant Microbe Interact. 2014;27:1379–1389. doi: 10.1094/MPMI-06-14-0190-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.