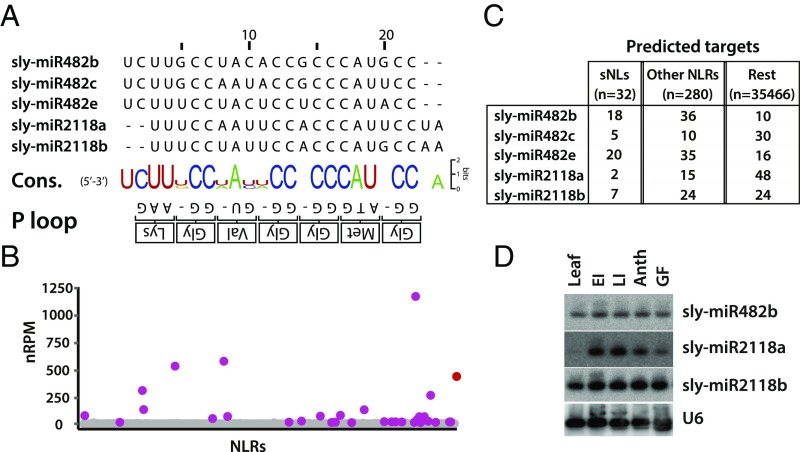
Fig. 1.
The miR482/2118 family in tomato. (A) Nucleotide sequence alignment of mature miR482/2118 members in tomato. The consensus sequence of each position in the alignment vs. the P-loop motif is shown at the bottom. (B) Dot plot representing the sum of 21-nt sRNA nRPM aligning to an individual NLR. A total of 32 NLRs presented >10 nRPM counts and were defined as sNLs (purple). TAS5 (red) is added as a reference. (C) Summary of target prediction of all miR482/2118 members. (D) RNA gel blot analysis of tomato miR482/2118 members in various tissues of plant development. Column 4 shows the same blot hybridized with U6 as a loading control. Anth, anthesis; EI, early influorescence; GF, green fruit; LI, late influorescence.