Significance
Sleep disturbances are common in stress-related disorders but the nature of these sleep disturbances and how they relate to changes in the stress hormone corticosterone and changes in gene expression remained unknown. Here we demonstrate that in response to chronic mild stress, rapid–eye-movement sleep (REMS), a sleep state involved in emotion regulation and fear conditioning, changed first and more so than any other measured sleep characteristic. Transcriptomic profiles related to REMS continuity and theta oscillations overlapped with those for corticosterone, as well as with predictors for anhedonia, and were enriched for apoptotic pathways. These data highlight the central role of REMS in response to stress and warrant further investigation into REMS’s involvement in stress-related mental health disorders.
Keywords: depression, anhedonia, EEG theta power, machine learning, transcriptome
Abstract
One of sleep’s putative functions is mediation of adaptation to waking experiences. Chronic stress is a common waking experience; however, which specific aspect of sleep is most responsive, and how sleep changes relate to behavioral disturbances and molecular correlates remain unknown. We quantified sleep, physical, endocrine, and behavioral variables, as well as the brain and blood transcriptome in mice exposed to 9 weeks of unpredictable chronic mild stress (UCMS). Comparing 46 phenotypic variables revealed that rapid–eye-movement sleep (REMS), corticosterone regulation, and coat state were most responsive to UCMS. REMS theta oscillations were enhanced, whereas delta oscillations in non-REMS were unaffected. Transcripts affected by UCMS in the prefrontal cortex, hippocampus, hypothalamus, and blood were associated with inflammatory and immune responses. A machine-learning approach controlling for unspecific UCMS effects identified transcriptomic predictor sets for REMS parameters that were enriched in 193 pathways, including some involved in stem cells, immune response, and apoptosis and survival. Only three pathways were enriched in predictor sets for non-REMS. Transcriptomic predictor sets for variation in REMS continuity and theta activity shared many pathways with corticosterone regulation, in particular pathways implicated in apoptosis and survival, including mitochondrial apoptotic machinery. Predictor sets for REMS and anhedonia shared pathways involved in oxidative stress, cell proliferation, and apoptosis. These data identify REMS as a core and early element of the response to chronic stress, and identify apoptosis and survival pathways as a putative mechanism by which REMS may mediate the response to stressful waking experiences.
Sleep is assumed to contribute to recovery from the wear and tear of wakefulness and to mediate adaptation to the waking experience, be it through memory consolidation or processing of emotional experiences, such as those associated with stressful events (1). Chronic stress is the most significant predictor of mood disorders (2) and major depressive disorder is anticipated to be the leading cause of disease burden by 2030 (3), while the true global burden of stress-related mental diseases might be largely underestimated (4). In animals, chronic stress leads to profound physiological changes, such as hypothalamic–pituitary–adrenal (HPA) axis regulation of corticosterone, neurogenesis, synaptic plasticity, and gene expression (5, 6). Chronic stress also leads to a plethora of behavioral disturbances, including depressive-like behavior, decreased responsiveness to rewards akin to anhedonia, a core symptom of depression, and sleep alterations (1, 7). The effects of chronic stress on sleep in rodents have been studied by applying physical, social, and environmental stressors. Several of these studies documented alterations in rapid–eye-movement sleep (REMS) and sleep continuity (8–11), while others reported changes in non-REM sleep (NREMS) and electroencephalogram (EEG) slow wave (delta) activity (12). In humans, chronic stress, alterations in the HPA axis regulating cortisol, and sleep disturbances have been associated with mood disorders (13, 14). However, the nature of sleep disturbances in major depression continues to be discussed, with some studies highlighting changes in NREMS (15, 16), and others REMS and sleep continuity (14, 17). Unresolved questions are how the various physiological and behavioral consequences of chronic stress interrelate and whether specific changes in sleep are early and core symptoms contributing to adaptation to chronic stress.
Stress triggers changes in gene expression in the brain and these transcriptome responses have been shown to be highly tissue/brain region-specific (6). Most studies have focused on the hippocampus and prefrontal cortex, identifying differential expression of genes related to inflammation, immune response, and neurogenesis (18–20). While transcriptomic changes underlying neuroplastic adaptation to chronic stress have been extensively studied in the brain, very few animal studies have investigated the transcriptome response to stress in blood (21, 22). This is of interest in the context of translational studies because blood transcriptomic signatures of depression and treatment response have been identified in humans (23–25). Finally, the extent to which sleep and other behavioral and endocrine alterations in response to stress are related to changes in the transcriptome has not yet been comprehensively quantified.
Here, exposure to chronic stress was achieved using the well-validated unpredictable chronic mild stress (UCMS) paradigm in mice (7). UCMS elicits a broad range of physiological and ethological changes that are consistent with symptoms of major depressive disorder, and predicts the efficacy of antidepressant treatments (7, 26). This ethological “model” has been recognized for its high-translational potential in the context of stress-related disorders (26–28). The aims of the current study were: (i) to comprehensively characterize chronic stress-induced changes in REMS and NREMS, corticosterone, and behavioral variables, as well as the transcriptome in three stress- and sleep-related brain regions (hippocampus, prefrontal cortex, hypothalamus) and blood; and (ii) to investigate the interrelationship of these responses using machine learning and other robust statistical approaches.
Results
Stress-Induced Physical, Neuroendocrine, and Behavioral Disturbances.
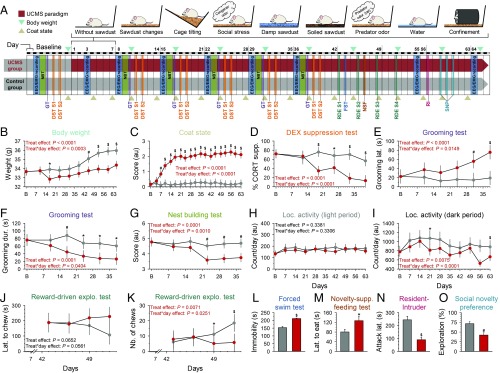
We assessed the impact of the repeated exposure to an unpredictable stressful waking experience on a number of physiological and behavioral variables during the 9-wk protocol (Fig. 1A). Chronic mild stress significantly altered body weight and worsened coat state, an index of reduced grooming behavior (Fig. 1 B and C). Corticosterone regulation was compromised in the UCMS group, consistent with blunted HPA axis negative feedback (Fig. 1D). The dexamethasone (DEX)-induced corticosterone suppression results are not explained by handling and injection because the response to saline injection was not different between groups (P = 0.657) (SI Appendix, Fig. S1). Self-care behavior was reduced, as reflected by increased grooming latency and decreased grooming duration (Fig. 1 E and F). Quality of nest building, indicative of motivation, was also reduced in the UCMS group (Fig. 1G). Moreover, UCMS suppressed the progressive increase of consumption of a palatable stimulus, indicative of anhedonia (Fig. 1 J and K). Immobility during the forced swim test was increased (Fig. 1L), as was anxiety-like behavior (Fig. 1M). Social disturbances were observed with increased aggressive behavior (i.e., decrease of attack latency and increased number of attacks) (Fig. 1N and Dataset S1), and decreased social preference for the novel congener (Fig. 1O). Exposure to UCMS reduced the weekly averaged locomotor activity during the dark (active) phase of the light–dark cycle, while activity remained unaffected during the light phase (Fig. 1 H and I and SI Appendix, Fig. S2). The lower locomotor activity of UCMS-subjected mice was also observed on stress-free days (i.e., during sleep recordings and the nest building test; P < 0.0001), suggesting a persistent effect of stress even when no stressor is applied.
Fig. 1.
UCMS protocol and physical, corticosterone regulation, and behavioral alterations. (A) Overview of the protocol. Mice were randomly assigned to the control (gray) or the UCMS (red) group. (B) Body weight, (C) coat state, (D) HPA axis negative feedback [dexamethasone (DEX) suppression test; DST; n = 5–7 per group], (E and F) self-centered behavior (grooming test; GT), (G) motivation (nest building test; NBT), and (H and I) locomotor activity, were measured at baseline and during the 9-wk UCMS. From day 43, several behavioral domains were evaluated, including (J and K) anhedonia-like (reward-driven exploratory test assessing the motivation to collect a palatable stimulus; latency and number of chews), (L) despair (depressive-like) behavior assessed by increased immobility in the forced swim test (FST; n = 8 per group), (M) anxiety-like behavior evaluated by increased latency to eat the food pellet in the novelty-suppressed feeding test (NSF), (N) aggressiveness identified by shorter attack latency in the resident-intruder (RI) test, and (O) social disturbances reflected by reduced time spent with the unfamiliar conspecific in the UCMS group (social novelty preference test; SNP). Data are shown for n = 9 per group (unless specified otherwise), as LSmean ± 95% CIs, except for (L–O) mean ± SEM; *P < 0.05, #P < 0.01, $P < 0.001 (post hoc comparisons for significant “treatment” × “day” interaction in general linear mixed model, or significant t test for nonrepeated measures). For detailed statistics, see Dataset S1. S, session.
Impact of 9-wk UCMS on Sleep.
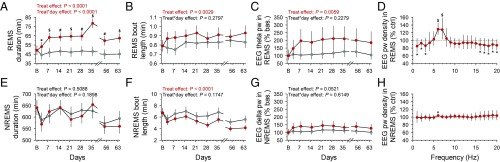
Twenty-four hour REMS duration increased significantly during UCMS (Fig. 2A). In contrast, 24-h total sleep time (TST) and NREMS duration were not significantly altered (TST: treatment effect P = 0.4727; interaction “treatment” × “day”: P = 0.0993) (Fig. 2E). Expressed as a percentage of TST, REMS was increased, and these changes were observed both during the light and dark phases (SI Appendix, Fig. S3 C and D). Chronic mild stress also induced an increase in REMS continuity, with increased duration of REMS episodes (Fig. 2B) despite increased number of REMS episodes during both the light and dark phases (SI Appendix, Fig. S3 G, H, K, and L). In contrast, NREMS became more fragmented with an increased number of episodes of shorter duration (Fig. 2F and SI Appendix, Fig. S3 E, F, I, and J).
Fig. 2.
Time course of UCMS-induced alterations on sleep and the EEG. (A) Duration of REMS per 24 h. (B) Duration of REMS episodes per 24 h. (C) EEG theta power density (6–9 Hz) in REMS during the 12-h light phase expressed as the percentage of theta power in baseline. (D) Relative EEG power spectra in REMS during the 12-h light phase (averaged spectra of all EEG recording sessions during the 9-wk UCMS protocol). (E) Duration of NREMS per 24 h. (F) Duration of NREMS episodes per 24 h. (G) EEG delta power density (1–4.5 Hz) during the 12-h light phase expressed as the percentage of delta power in baseline. (H) Relative EEG power spectra in NREMS (averaged spectra of all EEG recording sessions during the 9-wk UCMS protocol). Data are LSmeans ± 95% CI (controls: gray; UCMS: red; n = 8 per group); *P < 0.05, #P < 0.01, $P < 0.001 (post hoc comparisons for significant “treatment” × “day” interaction, except for D and H: effect of “treatment” in general linear mixed model). For detailed statistics, see Dataset S1.
Quantitative EEG analysis, using baseline measurements as a covariate to control for individual differences in the EEG power spectra, showed that theta activity, an EEG hallmark of REMS, was increased in the light (Fig. 2C) and dark phases (SI Appendix, Fig. S3 O and P). In contrast, NREMS delta activity was not affected by UCMS (Fig. 2G and SI Appendix, Fig. S3 M and N). Computation of relative EEG power spectra showed that changes in REMS were indeed mainly observed in the theta range, although some reduced activity in lower and higher frequencies was detected (Fig. 2D and SI Appendix, Fig. S4A). The increase in theta is not directly related to the duration of REMS bouts because power is a density measure that does not necessarily increase with bout duration. To further explore this issue, we compared theta power in long and short REMS bouts and nevertheless found that theta power is higher in long REMS bouts than in short REMS bouts in both UCMS and control groups (Dataset S1). We then compared theta power associated with long and short REMS bout lengths in light and dark periods between control and UCMS-subjected mice. We found that in both short and long REMS bouts, theta power was higher in the UCMS group, except for short REMS bouts in the light phase (Dataset S1).
In contrast to REMS, only minor changes were observed in the relative NREMS EEG power spectra (Fig. 2H and SI Appendix, Fig. S4B).
Temporal Associations of Phenotypic Alterations.
The changes in 24-h REMS duration, and other measures of sleep duration across 24 h or during the light phase, were observed as early as day 3 of the UCMS protocol (Fig. 2A and SI Appendix, Figs. S3 A–D and S5). Degradation of coat state occurred from day 7, while differences in body weight, impairment of corticosterone regulation, self-centered behavior, and motivation appeared in weeks 3–4 (Fig. 1 B–G and SI Appendix, Fig. S5). Locomotor activity in the dark period was reduced in the UCMS group during the last 3 wk of the 9-wk protocol (SI Appendix, Fig. S2 B–E).
Effect Size and Stability of Chronic Stress Effects Across Phenotypes.
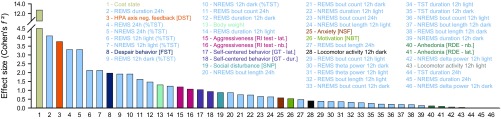
The size of the effects of UCMS varied considerably across dependent measures, with the largest effect sizes observed for coat state, 24-h REMS duration, corticosterone regulation, and 24-h REMS expressed as percentage of TST (Fig. 3). Overall, most REMS and NREMS variables, including the number and length of sleep episodes, displayed a large (Cohen’s f2 > 0.4) or medium effect size (Cohen’s f2 > 0.25) (Fig. 3). Across behaviors, effect sizes of UCMS were large for despair behavior, aggression, self-centered behavior, social disturbances, anxiety-like behavior, and motivation. The impact of UCMS on 24-h TST, 24-h NREMS duration, and EEG delta power was small (Fig. 3). In addition, to assess to which extent UCMS-induced changes were stable within individuals, intraclass correlation (ICC) coefficients were computed for all dependent variables. ICCs ranged between 0.67 and 0.997 for body weight, locomotor activity, REMS EEG theta power, and NREMS delta power, suggesting that the response to UCMS is highly stable (i.e., ICC > 0.61 benchmarks defined by ref. 29) within individuals. Coat state, as well as REMS and NREMS expressed as a percentage of TST for 24-h, showed moderate trait stability (ICC = 0.5240 and 0.4671, respectively). Corticosterone regulation displayed a slight stability (ICC = 0.0066) (Dataset S1).
Fig. 3.
Effect size of UCMS-induced physical, behavioral, neuroendocrine, and sleep alterations. Effect sizes of repeated (Cohen’s f2) and nonrepeated measures (Cohen’s d were converted to Cohen’s f2 using the following formula: Cohen’s d = 2 × Cohen’s f2), with large effect size: >0.40; medium: 0.25–0.40; small: 0.10–0.25. Bar colors correspond to those displayed in Fig. 1A and SI Appendix, Fig. S5 for all measured phenotypes. For values, see Dataset S1.
Bivariate Associations Between Phenotypes.
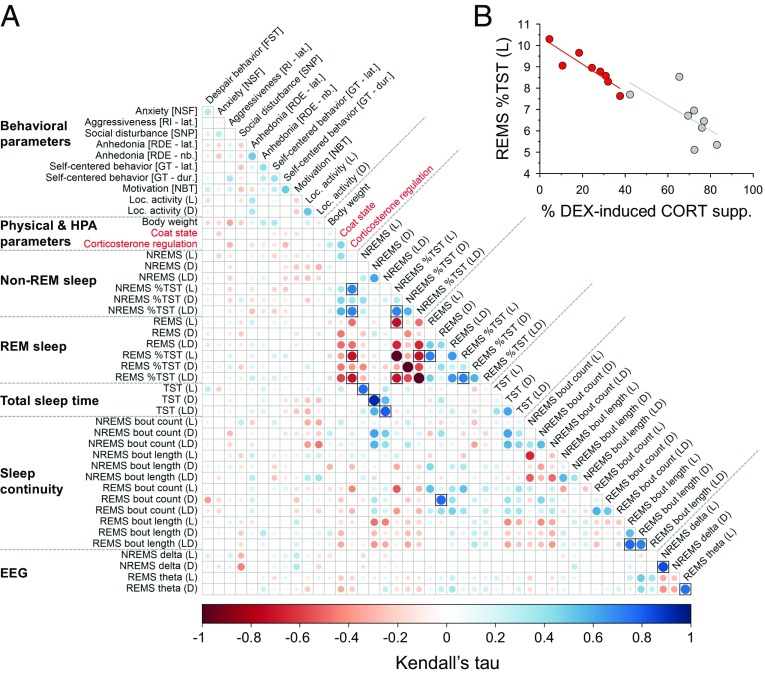
To assess the strength of associations between measured variables, we computed Kendall’s partial correlations between pairs of symptoms induced by chronic mild stress. We controlled for the effect of “group” (i.e., control vs. UCMS) to identify bivariate associations at the level of the individual independent of “unspecific” group effects. The increased percentage of REMS per TST (during the light phase and for 24 h) correlated negatively with DEX suppression; that is, more REMS was associated with the impairment of corticosterone regulation [τ = 0.72, nominal P value (Pnom) = 0.00034, false-discovery rate (FDR)-adjusted P value (Padj) = 0.0252 and τ = 0.71, Pnom = 0.00037, Padj = 0.0211, respectively] (Fig. 4 A and B). These associations were of a large effect size defined by τ > 0.25 (30). Other large effect-size associations were observed; however, they reflected trivial relationships among dependent sleep variables (e.g., percentage of REMS and NREMS per TST).
Fig. 4.
Bivariate associations for physical, behavioral, neuroendocrine, and sleep alterations. (A) Kendall’s partial correlation between pairs of phenotypes (i.e., after removing the effect of the experimental groups). The phenotypes (the averaged last three measurements were used for repeated measures) were ordered according to their phenotypic categories. Correlations were considered significant at an FDR < 0.05 (Padj; symbolized by black-framed square), computed with the Benjamini–Hochberg procedure for multiple testing correction. For detailed statistics, see Dataset S2. (B) Example of a correlation from (A) illustrated for percentage of REMS per TST during the light (L) phase and impairment of the corticosterone regulation (τ = 0.72; Pnom = 0.00034, Padj= 0.0197; n = 8 animals per group; gray: control mice, red: UCMS-subjected animals). DEX supp., dexamethasone suppression.
Effects of Chronic Stress on the Transcriptome.
To gain insight into the molecular mechanisms underlying the phenotypes induced by UCMS, we performed RNA sequencing on three brain regions and whole-blood samples collected at the end of the UCMS paradigm.
Differential gene expression and functional enrichment.
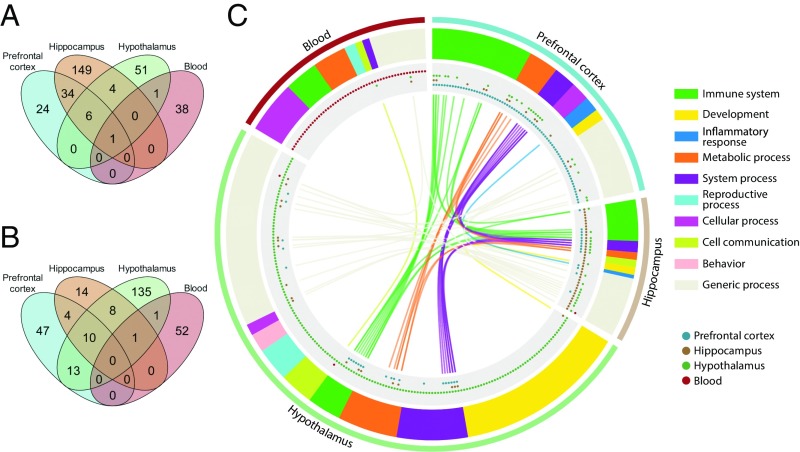
We first performed differential expression analysis between the UCMS and control groups. The number of differentially expressed genes (DEGs) was relatively small (range across the three brain regions and blood: 40–194) and the number of up-regulated genes was larger than the number of down-regulated genes in all tissues (Dataset S3). The fold-changes were relatively small (range of log2-transformed fold-change: −1.65 to 1.18) (Dataset S3). The comparison of transcriptomic responses in the four tissues showed a robust overlap of DEGs between the prefrontal cortex and the hippocampus, while the commonalities between other tissues were weaker (Fig. 5A; for identity of these overlapping DEGs, see SI Appendix, Fig. S6 and Dataset S3). The three brain regions had only six common DEGs, encoding hemoglobin subunits (Hba-a1, Hba-a2, Hbb-b1, Hbb-bs), an erythroid-specific mitochondrially located enzyme (Alas2), as well as the noncoding RNA Rprl2. Only one DEG, the predicted gene Gm8221 (apolipoprotein L 7c pseudogene), was common to all four tissues and was among the most down-regulated DEGs in all tissues (Fig. 5A and Dataset S3). At the individual transcript level, a literature search revealed that numerous DEGs in all four tissues had been previously reported to be associated with sleep and circadian rhythms (prefrontal cortex: 35.1%; hippocampus: 18.7%; hypothalamus: 21.1%; blood: 17.1%), stress (prefrontal cortex: 40.5%; hippocampus: 35.2%; hypothalamus: 50.9%; blood: 20%), neuropsychiatric symptoms (prefrontal cortex: 37.8%; hippocampus: 20.9%; hypothalamus: 29.8%; blood: 25.7%), mood disorders (prefrontal cortex: 16.2%; hippocampus: 8.8%; hypothalamus: 19.3%; blood: 2.9%), or neurodegenerative diseases, such as Alzheimer’s and Parkinson’s diseases (prefrontal cortex: 37.8%; hippocampus: 30.8%; hypothalamus: 36.8%; blood: 17.1%) (see SI Appendix, Fig. S6A and Dataset S3 for references). In addition, several DEGs in the prefrontal cortex (e.g., S100a8, S100a9, Lbp, Tgtp2), hippocampus (e.g., Inava, Lbp, Rsad2, Pla2g5, F5, Vegdf, Cd24a, Tgtp1, Cast, Lst1), and blood (e.g., Clec4n, Chil3, Reg3g, Bpifa1) play a key role in the immune system, inflammation, and the vascular system. Both the hippocampus (Glra3, Ptgdr, Pmch, Oprk1, Kcne2, Gpr6) and hypothalamus (Slc6a3, Slc5a7, Chat) showed several DEGs involved in neural transmission, including the down-regulation of neuropeptide-encoding genes implicated in adaptation to stress and social behavior in the hypothalamus (i.e., Ucn3, Avp, Oxt, Vip). Some of the most up-regulated DEGs in blood are involved with DNA damage response (i.e., Mnd1, E2f7), while others have been previously associated with sleep deprivation or fragmentation (Fads3, Gm6166, Spp1, Hspa1a, Hspa1b, Scgb3a1) (SI Appendix, Fig. S6A and Dataset S3).
Fig. 5.
Characterization and functional enrichment of genes differentially expressed following chronic mild stress. Overlap of (A) DEGs and (B) significantly enriched GO biological processes for DEGs in the prefrontal cortex, hippocampus, hypothalamus, and whole blood. (C) GO biological processes associated with DEGs. Outer track: tissue; second track: overarching themes associated with GO processes; third track: tissues in which GO processes were found; inner track: overlap of processes, colors corresponding to overarching theme. n = 8 per group for brain regions; n = 7 controls vs. n = 9 UCMS group for blood. Enrichment analyses were performed using MetaCore and significance was set at Padj < 0.05. Information is available in tabular format (Datasets S3 and S4).
To further characterize the effects of the 9-wk UCMS, we performed functional enrichment analysis using Gene Ontology (GO) processes and canonical pathway maps. The hypothalamus showed the largest number of enriched GO processes (n = 168) compared with the prefrontal cortex (n = 74), hippocampus (n = 37), and blood (n = 54). Ten processes were shared by the three brain regions (Fig. 5B). These included processes associated with the immune system (i.e., erythrocyte development and differentiation), circulatory system processes (e.g., regulation of blood pressure), and metabolic processes (e.g., oxygen transport, hydrogen peroxide metabolic process) (Fig. 5C; for detailed identity of the GO processes, see Dataset S4). In contrast, only two enriched GO processes were common to blood and brain regions. Response to stress was common to the blood, hypothalamus, and hippocampus, while regulation of receptor activity was shared by blood and hypothalamus (Fig. 5C and Dataset S4).
GO biological processes in the hypothalamus were involved in developmental processes (e.g., cell fate commitment), nervous system processes (e.g., regulation of sensory perception), immune system (e.g., regulation of C-C chemokine binding, myeloid cell homeostasis), cell communication (e.g., G protein-coupled receptor signaling pathway), and behavior (grooming and aggressive behaviors) (Fig. 5C and Dataset S4). One enriched pathway involved in protein folding and maturation (i.e., posttranslational processing of neuroendocrine peptides) was observed (SI Appendix, Fig. S7 and Dataset S4). In the prefrontal cortex and hippocampus, functional enrichment analysis identified 37 processes associated with inflammatory and immune response (some of which were shared; e.g., response to IFN-β; leukocyte migration involved in inflammatory response) among others (Fig. 5C and Dataset S4). Enriched pathways evoked by chronic stress were involved in transcription and development; however, none were significant in the hippocampus after FDR adjustment (SI Appendix, Fig. S7 and Dataset S4). In blood, functional enrichment also identified biological processes involved in immune and inflammatory response (e.g., regulation of cytokine production) and signaling pathways (e.g., nitric oxide-mediated signal transduction, TNF-mediated signaling pathways). In addition, 10 processes were associated with RNA cleavage and the unfolded protein response (Fig. 5C and Dataset S4).
Bivariate correlations between molecular consequences of chronic stress and phenotypic disturbances.
To identify associations between DEGs and phenotypic alterations induced by UCMS, we performed bivariate analyses, computing Kendall’s partial correlations in which the effect of “group” (i.e., control vs. UCMS) was controlled for, for all physical, neuroendocrine, behavioral, and sleep variables and DEGs per tissue. We observed that 26.3% (821 of 3,120), 25.9% (2,413 of 9,312), 20.7% (626 of 3,024), and 29.5% (566 of 1,920) of the associations between DEGs and stress-induced symptoms exhibited a correlation of large effect size (i.e., τ > 0.25) in the prefrontal cortex, the hippocampus, the hypothalamus, and the blood, respectively (Dataset S5). In the hippocampus, Inava, encoding the innate immunity activator, was associated with REMS bout length in the light and dark periods (τ = 0.49 and 0.7, respectively). Ucn3 and Vip were associated with REMS bout length in the light period (hypothalamus; τ = −0.41 and 0.25). However, no correlation remained significant after FDR adjustment.
Selection of transcriptomic predictor sets associated with phenotypes using a penalized regression approach.
While univariate approaches provide some insights into the associations between transcripts and other physiological and behavioral variables, they nevertheless suffer from the multiplicity problem. In addition, they are not necessarily best suited to identify sets of transcripts that predict specific complex phenotypes. Thus, we applied elastic-net learning, a multivariate approach based on a generalized linear model using penalized regression, to identify sets of features predicting specific phenotypes. We performed this analysis using all transcripts identified by RNA sequencing, that is, not just the DEGs, focusing on sleep variables and some variables associated with stress and mood disorders. We aimed to identify transcriptomic features that were specifically associated with sleep and behavioral variables both within the control and UCMS group (i.e., at the level of the individual). To accomplish this, “unspecific group” effects (i.e., control vs. UCMS) on these variables need to be removed from the analysis. We therefore applied normalization procedures to control for group effects (Materials and Methods). The features that associate with behavioral variables, as identified by elastic-net after application of the normalization procedures, indeed contained very few transcripts (30 of 1,595) identified by the group-level analysis (DEGs; see previous section). This demonstrates that this approach yields information that is different from the DEG approach. The number of features in the various identified predictor sets was overall small and varied between variables and across tissues (range: 1–333) (Dataset S6). To gain insights into molecular mechanisms associated with a given sleep or behavioral variable and to contrast biological correlates of the sleep and other variables, we then performed functional enrichment analysis of predictor sets focusing on pathway maps.
REMS and NREMS.
The size of the predictor sets for REMS and NREMS parameters was similar for sleep duration and continuity (493 and 464, respectively), but few significantly overlapped (n = 73) (Dataset S7). Common predictors were seen in the prefrontal cortex, primarily between REMS continuity and NREMS duration (n = 29) and continuity (n = 39), as well as in the hippocampus between REMS bout count and NREMS duration (n = 3) and bout count (n = 2) (Dataset S7). Looking individually at transcriptomic features for REMS and NREMS variables, several included transcripts involved in neural transmission, sleep, and circadian rhythms (Dataset S8). Six predictors for REMS variables play a key role in the regulation of NF-κB signaling, while 26 predictors of NREMS were associated with mitochondrial function (Dataset S8).
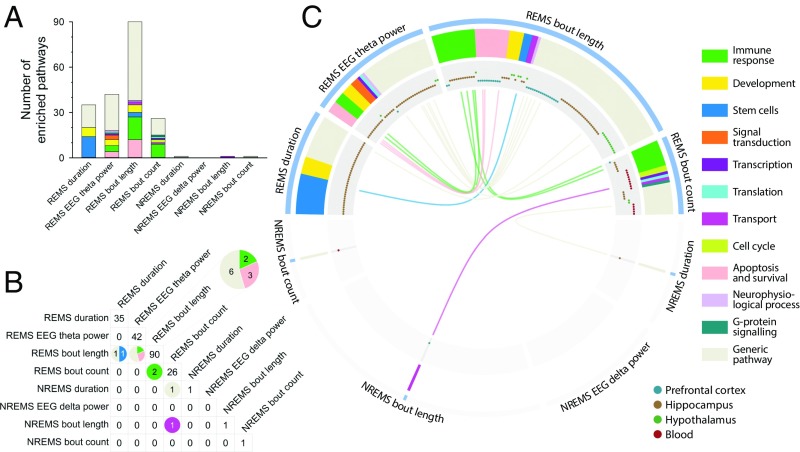
Whereas REMS predictor sets were significantly enriched for many canonical pathways (n = 193), only three pathways were identified in the NREMS predictor sets (Fig. 6 A–C and Dataset S9). Enriched pathways associated with predictors of REMS duration and REMS theta power were primarily observed in the hippocampus (n = 35 and 42, respectively), while pathways associated with predictors of REMS bout length (n = 90) and REMS bout count (n = 26) were primarily enriched in the prefrontal cortex (n = 30), hippocampus (n = 43) and hypothalamus (n = 17), and in the hippocampus (n = 9) and blood (n = 14), respectively (Fig. 6C). REMS duration displayed several enriched pathways associated with stem cells (n = 14) and development (n = 6) (Fig. 6 and Dataset S9). Across REMS theta power, REMS continuity and tissues, 24 pathways were involved in the immune response, including various IL, IFN, and Toll-like receptor signaling pathways. Remarkably, 11 enriched pathways were common to REMS theta power and REMS bout length, with eight of them associated with apoptosis and cell survival, such as the TNF receptor (TNFR)-1 signaling pathway, the role of inhibitor of apoptosis proteins (IAP), the endoplasmic reticulum (ER) stress response pathway (five of these pathways are listed under the theme “generic pathway” in Fig. 6). Of the three significantly enriched pathways identified for NREMS predictor sets, two pathways (i.e., Ras-related nuclear protein regulation, fructose pathway) overlapped with REMS continuity variables (Fig. 6 B and C).
Fig. 6.
Enriched pathways in transcriptomic predictor sets of sleep variables. (A) Number of enriched pathways associated with REMS and NREMS variables. Colors correspond to functional themes identified by the “Pathway Maps” tool in MetaCore. (B) Number of overlapping pathways between REMS and/or NREMS variables. Colors correspond to the functional themes of pathways. (C) Enriched REMS- (Upper) and NREMS- (Lower) associated pathways. Outer track: phenotypes; second track: functional themes of pathways; third track: tissues in which pathways were found to be significantly enriched; inner track: overlaps between pathways are illustrated with color of functional themes. All depicted pathways were significant at Padj < 0.05; n = 8 per group for brain regions; n = 7 controls vs. n = 9 UCMS group for blood. Lists of enriched pathways are available in tabular format (Dataset S9).
Corticosterone regulation, anhedonia, and despair behavior.
Some transcripts in the cortical predictor set for corticosterone regulation were associated with neural transmission and psychiatric conditions (Dataset S8). Several sleep- and circadian-related genes were seen in feature sets of despair behavior and anhedonia (Dataset S8). Functional analysis of predictors for corticosterone regulation identified 40 enriched pathways in the hippocampus. These were primarily associated with apoptosis and cell survival (n = 12, including 4 listed under the “generic” theme) (Dataset S9), stem cells (n = 5), immune response (n = 3), development (n = 3), as well as several generic metabolic and signaling pathways (SI Appendix, Fig. S8). Predictor sets of anhedonia were enriched in several pathways involved in development (n = 12) and stem cell processes (n = 7), as well as transcription (n = 4) and generic pathways (SI Appendix, Fig. S8). Nine enriched pathways were found in predictor sets for despair behavior, primarily in the hippocampus (n = 7) and included circadian rhythm process (SI Appendix, Fig. S8 and Dataset S9).
Pathways shared between REMS, corticosterone regulation, and anhedonia.
More than one-third of the 40 pathways associated with corticosterone regulation (37.5%) overlapped with pathways for REMS bout length or for EEG theta activity in the prefrontal cortex and hippocampus, respectively (SI Appendix, Fig. S8). Thirteen of the 15 common pathways were associated with apoptosis and cell survival (Table 1). They included apoptotic pathways involved in the extrinsic death receptor pathway (e.g., TNFR-1 signaling, FAS signaling cascade, and apoptotic TNF-family pathways) and the intrinsic mitochondrial pathway (role of IAPs in apoptosis; regulation of apoptosis by mitochondrial proteins) (Table 1 and SI Appendix, Fig. S8). In addition, REMS variables, and in particular REMS bout length and EEG theta activity, also shared seven pathways with anhedonia. These included pathways involved in apoptosis and survival and response to oxidative stress (Table 1). Finally, no overlap was observed between pathways associated with NREMS variables, corticosterone regulation, and any of the investigated behavioral variables (SI Appendix, Fig. S9).
Table 1.
Enriched pathways shared by REM sleep and corticosterone regulation, or REM sleep and anhedonia
| REMS variables | Overlapping pathways | Apoptosis and survival related pathway |
| Corticosterone regulation | ||
| Bout length | TNFR1 signaling pathway* (PFC) | √ |
| Bout length | Role of IAP-proteins in apoptosis* (PFC) | √ |
| Bout length | Granzyme B signaling* (PFC) | √ |
| Bout length | Regulation of apoptosis by mitochondrial proteins* (PFC) | √ |
| Bout length | Cytoplasmic/mitochondrial transport of proapoptotic proteins Bid, Bmf, and Bim* (PFC) | √ |
| Bout length | Apoptotic TNF-family pathways* (PFC) | √ |
| Bout length | FAS signaling cascades* (PFC) | √ |
| Bout length | Endothelial differentiation during embryonic development† (HYP) | |
| Bout length | Role of Apo-2L(TNFSF10) in Prostate Cancer cell apoptosis‡ (PFC) | √ |
| Bout length | Resistance of melanoma cells to Apo-2L(TNFSF10)-induced apoptosis‡ (PFC) | √ |
| Bout length | Inhibition of apoptosis in gastric cancer‡ (PFC) | √ |
| Bout length | Neutrophil resistance to apoptosis in chronic obstructive pulmonary disease and proresolving impact of lipid mediators‡ (PFC) | √ |
| Bout length | Apo-2L(TNFSF10)-induced apoptosis in melanoma‡ (PFC) | √ |
| Bout length | Apoptotic pathways and resistance to apoptosis in lung cancer cells‡ (PFC) | √ |
| Duration | Endothelial differentiation during embryonic development† (HIP) | |
| Theta power | Role of IAP-proteins in apoptosis* (HIP) | √ |
| Theta power | TNFR-1 signaling pathway* (HIP) | √ |
| Theta power | IL-5 signaling via JAK/STAT§ (HIP) | |
| Theta power | Role of Apo-2L(TNFSF10) in prostate cancer cell apoptosis‡ (HIP) | √ |
| Theta power | Inhibition of apoptosis in gastric cancer‡ (HIP) | √ |
| Theta power | Apoptotic pathways and resistance to apoptosis in lung cancer cells‡ (HIP) | √ |
| Anhedonia | ||
| Bout count | Metabolism in pancreatic cancer cells‡ (BLO) | |
| Bout length | Cytoplasmic/mitochondrial transport of proapoptotic proteins Bid, Bmf, and Bim* (PFC) | √ |
| Bout length | Glucocorticoids-mediated inhibition of proconstrictory and proinflammatory signaling in airway smooth muscle cells‡ (HIP) | |
| Bout length | Memory CD8+ T cells in allergic contact dermatitis‡ (HYP) | |
| Duration | EGF-induced proliferation of Type C cells in secondary proliferative zone of adult brain† (HIP) | |
| Theta power | Memory CD8+ T cells in allergic contact dermatitis‡ (HIP) | |
| Theta power | NRF2 regulation of oxidative stress response‡ (HIP) | |
Tissue in which enriched pathways associated with predictors of REMS variables is indicated in parenthesis (i.e., BLO, blood; HIP, hippocampus; HYP, hypothalamus; PFC, prefrontal cortex). The column “Apoptosis and survival related pathway” indicates whether a pathway was related to apoptosis and survival as some pathways listed under a generic theme in MetaCore are also implicated in apoptotic and antiapoptotic pathways. For each pathway, the functional theme to which it belongs to is listed according to the specified footnote.
Apoptosis and survival.
Stem cells.
Generic (according to MetaCore).
Immune response.
Discussion
REMS Enhancement, a Core Response to Chronic Mild Stress.
The UCMS paradigm induced changes in physical, behavioral, and neuroendocrine variables in accordance with previous reports (7, 20, 28). The simultaneous and longitudinal assessment of a wide range of variables allowed for a comparison of the magnitude of changes and the temporal emergence of these physical, behavioral, and neuroendocrine alterations. This approach demonstrated that increase in REMS variables (i.e., 24-h duration, bout length, bout count, EEG theta oscillations) exhibited not only large effect sizes but were also among the earliest responses induced by stress. The longitudinal assessment of sleep also demonstrated that REMS and NREMS respond differently to chronic stress. The increased continuity (i.e., bout length, bout count, and duration of REMS) and the increase in EEG theta activity during REMS, primarily reflecting hippocampal theta activity, imply that REMS is affected by UCMS in a positive manner. Of particular interest is the increase in both REMS bout duration and theta power, because in the rat it has been reported that theta power decreases in the course of a REM bout (31). In contrast, NREMS continuity was decreased and EEG delta power in NREMS was not affected. The changes observed in sleep and their effect sizes agree well with metaanalyses performed in clinical depressive populations, according to which effect sizes for REMS are larger than those for NREMS and sleep continuity (14, 17).
Transcripts and Associated Processes Affected by Chronic Mild Stress.
Transcriptome changes, assessed by differential expression and thus primarily reflecting effects of stress at the group level, were relatively small and most changes were observed in the hippocampus, which is consistent with previous reports (20, 21, 32). One tentative conclusion from these data are that effects of stress on sleep and changes in gene expression converge on the hippocampus (33), and an emerging question is whether REMS-related phenomena, such as EEG theta power, reflect or direct these hippocampal changes. On the other hand, our data also highlight that the cortex and hypothalamus are responsive to stress. In fact, the hypothalamus showed the largest number of enriched GO processes. Furthermore, many enriched biological processes were shared across brain regions. These include processes associated with inflammatory and immune responses, and parainflammation thus appears to be a common mechanism in the three brain regions investigated. This is in line with a recent framework emphasizing that inflammatory signals contribute to restore homeostasis (34) and agrees well with the emerging view that chronic stress and stress-related diseases, such as major depression, share inflammation as a common mediator (35, 36). Given the changes in DEX-induced corticosterone suppression, it may seem surprising that we did not observe changes in transcripts related to glucocorticoid or mineralocorticoid receptors in either the hippocampus or other brain regions. Our findings are, however, consistent with previous UCMS studies in which no change in their gene expression was observed (18, 20, 37).
Transcriptomic Predictors of Phenotypic Variation Identified Using Machine Learning.
Transcriptomic predictor sets were overall relatively small in accordance with a previous study (38). Hippocampal transcriptomic predictors of 24-h REMS duration were associated with pathways involving stem cells differentiation and hedgehog signaling. Inhibition of hedgehog signaling by glucocorticoid treatment has been shown to decrease hippocampal cell differentiation (39). In addition, several enriched pathways in apoptosis and cell survival were among the molecular signatures characterizing REMS continuity variable (bout length) and EEG theta power, in the cortex and hippocampus, respectively. The overlap between pathways for theta power and REMS bout length may point to common mechanisms underlying theta and bout length regulation. The identified predictors and related pathways specifically associated with REMS in the cortex, a brain region not necessarily implicated in the generation of REMS, may reflect an effector system by which REMS exerts its adaptive response to chronic stress. The ER stress-response pathway was common to REMS continuity and theta power. ER stress, which may lead to apoptosis (40), has been recently shown to be induced during social isolation in Drosophila (41). A number of circadian-related transcripts were identified as predictors of NREMS variables, as well as despair- and anhedonia-like behaviors. This is consistent with a recent study correlating UCMS-induced depressive-like behavior with circadian rhythm alterations in brain tissues (42), and the growing recognition that circadian rhythmicity may play a role in mood regulation (43, 44). It should be noted that while we observed changes in sleep, at the behavioral level circadian rhythmicity was not much affected, although the reduced activity during the dark period may be interpreted as a reduction in circadian amplitude. NREMS and REMS shared very few predictors, further emphasizing the contrast between these two sleep states observed at the electrophysiological level in this study. Furthermore, no overlap was observed between pathways associated with predictors for NREMS variables, corticosterone regulation, or behavioral phenotypes.
One aim of the present analyses was to investigate to what extent transcriptomic changes in the brain are reflected in the blood transcriptome. The results demonstrate that, at the level of individual DEGs or associated processes/pathways, there were no significant overlaps between brain and blood transcriptomic changes. However, even though the blood transcriptome may not be directly informative about changes in the brain, the elastic-net approach indicated that whole blood contains predictors of behavior (anhedonia) and REMS (bout count), which ultimately may be useful for biomarker development.
Close Associations Between REMS, Corticosterone Regulation, and Apoptotic Pathways.
One major theme emerging from the multilevel analyses is the robust effects of stress on REMS and the close link between changes in REMS and dysregulation of corticosterone. These data may be interpreted as evidence for a shared role of REMS and corticosterone within the context of “adaptation” to the waking experience. REMS has been proposed to play a central role in emotional processing and memory consolidation (45–48). A causal role for EEG theta activity during REMS was demonstrated in contextual and extinction memory consolidation in rodents and humans (49, 50). Furthermore, REMS is suppressed by most antidepressants (51) and some antidepressants interfere with the homeostatic control of REMS (52).
While REMS enhancement and alterations in the HPA axis negative-feedback regulation of corticosterone have been previously reported in preclinical studies of chronic stress or stress-vulnerable rodents (8, 9, 28), the present data demonstrate the close association between REMS% and corticosterone suppression (Fig. 4). This result, as well as previous findings in humans (53, 54), suggest that these phenomena share common causal mechanisms. Hypothalamic neuropeptides, such as vasoactive intestinal peptide (VIP), arginine vasopressin (AVP), and melanin-concentrating hormone (MCH), whose encoding genes were down-regulated in the present study, are potential candidates for orchestrating this association because they have been implicated in the regulation of REMS (55–58) and the HPA axis (59, 60). In humans, increased REMS (61) and HPA axis dysregulation (13) have been shown to correlate with remission and recovery in major depressive disorder.
Further evidence for the close association between REMS and corticosterone regulation emerged from the transcriptomic analyses. We identified 15 overlapping pathways between the corticosterone regulation and REMS continuity variables and theta power. These pathways were primarily involved in apoptosis and cell survival and included several members of TNF signaling, which triggers a broad spectrum of actions at the cellular level, including processes involved in the mitochondrial intrinsic pathway. Involvement of apoptotic pathways in depression and stress has been reported in recent human and animal studies. Blood transcriptomic studies in humans show that major depressive disorder and antidepressant response are associated with enrichment in apoptosis signaling processes and pathways (62, 63). Repetitive transcranial magnetic stimulation to treat depression counteract hippocampal neuronal apoptosis and HPA axis disturbances induced by UCMS in rats (64). In humans, chronic insufficient sleep, characterized by REMS alterations (65), is associated with apoptosis-related blood mRNA biomarkers (66), and sleep restriction in mice alters apoptotic pathway signaling (67).
We also identify shared molecular pathways underlying the interindividual variation in anhedonia, a core symptom of depression, with REMS continuity and theta power in response to chronic stress. They included pathways involved in oxidative stress and apoptosis with a pathway involved in the transport of proapoptotic proteins linking the Jun amino-terminal kinases (JNK) signal transduction pathway and the mitochondrial apoptotic machinery (68). A causal role for mitochondrial genes was recently proposed as part of the processes in the striatum, linking REMS and a stress-induced anxiety-like phenotype (38), and the contribution of mitochondrial dysfunction in major depression is emerging (69).
Considering the link between REMS, cell proliferation, and apoptosis (70, 71), the severe alterations of hippocampal neuronal plasticity and HPA axis functioning in mood disorders (72), and the strong responsiveness of the hippocampus to stress hormones (73), our results linking REMS continuity, theta oscillations, corticosterone regulation, and cell apoptosis as well as the shared pathways between REMS and anhedonia, shed a new light on the pathological framework of stress-related conditions.
Limitations.
Limitations of this study include a relatively small sample size and the choice of setting the statistical significance at FDR-adjusted P < 0.05, which may have led to an underreporting of significant effects. Another limitation relates to experimental constraints, which precluded an assessment of the temporal association between behavioral phenotypes and transcriptomic changes. Nevertheless, the high intraindividual stability of many of the phenotypes indicates that the observed transcriptomic changes at the end of the experiment are relevant to the phenotypes throughout the UCMS. This study was only conducted in males. Hypotheses based on the present data may be tested in future studies in which sex differences in sleep disturbances and their underlying molecular mechanisms in the context of chronic stress could be investigated (74).
Whether mice recover from the depressive-like phenotype after cessation of the UCMS was not assessed in this study. Other studies reported persistence of alterations at the transcriptome, metabolic, and behavioral levels for several days or weeks after the end of UCMS (75–78). How this pattern of recovery relates to changes in REMS has not yet been studied in detail.
Conclusion
This study in mice provides a comprehensive characterization of sleep changes induced by chronic stress, with REMS increase being the earliest marker of a stress response. Our data show that interindividual variation in REMS continuity and theta oscillations during REMS, and apoptosis processes including mitochondrial pathways, changes in corticosterone regulation, and anhedonia are interrelated. Alteration in corticosterone regulation and REMS have both been implicated in the response to emotional experiences. Given the prominence of REMS alterations in mood disorders and the herein-identified correlates of REMS, further study of the function of REMS parameters—such as its duration, continuity, and the theta oscillations during REMS—in the response to stress is warranted.
Materials and Methods
Animals.
Male BALB/cJ mice (n = 18; B&K Universal Ltd) underwent EEG/EMG surgery, as previously described (79) (see SI Appendix for details). After recovery, mice were randomly assigned to the control or UCMS group. Baseline data collection was performed, after which the 9-wk UCMS protocol started. Mice were daily subjected during the dark period to various socio-environmental low-intensity stressors according to an unpredictable schedule (27) (Fig. 1A and SI Appendix, Table S1). Experimental procedures were approved by the University of Surrey Animal Welfare and Ethical Review Body and were carried out in accordance with the United Kingdom Animals (Scientific Procedures) Act 1986.
Physical, Behavioral, and Corticosterone Regulation Assessments.
Body weight, coat state, self-centered behavior (grooming test), motivation (nest building test), anhedonia (reward-driven exploratory test), social preference (social novelty preference test), aggressiveness (resident-intruder test), anxiety (novelty-suppressed feeding test), and despair behavior (forced swim test) were assessed as previously described (27, 28, 80, 81) (see SI Appendix for details). The DEX suppression test was used to evaluate the HPA axis negative-feedback–regulated corticosterone (28) (see SI Appendix for details).
Sleep and Locomotor Activity.
No stressor was applied during the sleep recordings. The data analyzed consisted of 24-h recordings starting at dark onset. EEG power spectra were computed for consecutive 10-s epochs by a fast Fourier transform (see SI Appendix for details). Locomotor activity was measured as previously described (79). Averaged daily activity for the 12-h light and dark periods were analyzed per week.
Transcriptome Analysis.
Tissues (prefrontal cortex, hippocampus, hypothalamus, and whole blood) were collected 14–16 h after the last stress exposure. For details of RNA sequencing, see SI Appendix. Differential expression analysis (control vs. UCMS) was performed with the nonparametric Rank Product statistical method that is independent of interclass variability (82), using the R Bioconductor package RankProd. Significance was set at a proportion of false-positive (Ppfp) < 0.05. To robustly select relevant transcriptomic predictors, a form of penalized regression, referred to as elastic-net, was performed using the R package glmnet (83). Analyses were focused on sleep variables, three behavioral variables, and corticosterone regulation, the values of which were z-scored within group to identify associations with phenotypes independent of any “unspecific” group effect. We only report predictor sets for variables achieving a positive Pearson correlation r between observed and cross-validated prediction values >0.31. For functional annotation, lists of genes associated with a variable (e.g., REMS duration) were subjected to GO enrichment analyses (GO processes or Pathway maps) using MetaCore (Clarivate Analytics; https://portal.genego.com/; updated June 2018). Functional analyses were performed using the respective tissue-specific transcriptome, as identified by RNA sequencing, as background. Significant enrichment was defined by Pnom < 0.05 and Padj < 0.05.
Statistics.
Unless otherwise stated, data were analyzed with SAS 9.2 (SAS Institute). For repeated measures, data were analyzed as dependent variables in a general linear mixed model using PROC MIXED for ANOVA with group (treatment: UCMS vs. control) and time (day, treated as repeated measure with spatial power anisotropic variance-covariance matrix) as categorical explanatory variables with baseline as a covariate (no group effect was found at baseline for all measures). Post hoc multiple pair-wise comparisons (UCMS vs. control) were assessed using the ESTIMATE option of PROC MIXED. Output data are expressed as least-squares means (LSmeans) with 95% confidence intervals (CIs). For nonrepeated measures, PROC TTEST for pair-wise comparisons was used using Pooled or Satterthwaite methods for equal and unequal variances, respectively. Output data are expressed as mean ± SEM. Comparison of theta power in long and short REMS bouts, as well as comparison of locomotor activity during stress-free days in UCMS group and corresponding days in control group, were performed using PROC TTEST for repeated measures. Statistical effect sizes were calculated as Cohen’s f2 effect sizes (84). To assess the variability within experimental groups throughout time, ICCs were computed (29). Kendall’s partial correlations (Kendall’s τ coefficients), with experimental group as the controlled variable, were computed using PROC CORR for generating the phenotypic associations between output measures, while the FDRs using the Benjamini–Hochberg procedure for multiple testing correction were computed using the p.adjust function in R. Correlations were considered significant at Padj < 0.05. For repeated measures, the average of the last three measures was used for the calculation of the correlations. Despite some of the sleep variables conveying the same information (e.g., wake vs. TST), removing the duplicate variables did not alter the array of correlations reaching a significant Padj < 0.05 in the bivariate analyses. Some of these variables were therefore included in the figure for a comprehensive presentation of the data.
Supplementary Material
Acknowledgments
We thank Drs. Sig Johnsen and Jeewaka Mendis for advice and support with statistical analyses; Drs. Daan van der Veen and Bruno Martynhak for help with locomotor activity data analyses; Dr. Gillian Stenson for support during data acquisition and double-scoring of behavioral tasks; Drs. Hugh Marston and Dale Edgar for support; Dr. Lisiane Meira for helpful discussions; and Dr. Simon Archer for comments on the manuscript. This study was supported by a Lilly Innovation Fellowship Award (to M.N.) and conducted through an academic–industrial partnership between the Surrey Sleep Research Centre of the University of Surrey and Eli Lilly and Company. This paper was also supported by a Royal Society Wolfson Research Merit award (to D.-J.D.).
Footnotes
Conflict of interest statement: This study was supported by a Lilly Innovation Fellowship Award (to M.N.) and conducted through an academic–industrial partnership between the Surrey Sleep Research Centre of the University of Surrey and Eli Lilly and Company Ltd. K.A.W., A.P.M., K.M., N.L., and M.N. were full-time employees of Eli Lilly and Company Ltd. at the time of the study. D.-J.D. has received research funds and acted as consultant to Eli Lilly and other pharmaceutical companies. R.W.-S. has received research funding from Eli Lilly.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE125441).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816456116/-/DCSupplemental.
References
- 1.Sanford LD, Suchecki D, Meerlo P. Stress, arousal, and sleep. Curr Top Behav Neurosci. 2015;25:379–410. doi: 10.1007/7854_2014_314. [DOI] [PubMed] [Google Scholar]
- 2.Tafet GE, Nemeroff CB. The links between stress and depression: Psychoneuroendocrinological, genetic, and environmental interactions. J Neuropsychiatry Clin Neurosci. 2016;28:77–88. doi: 10.1176/appi.neuropsych.15030053. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization 2008 The Global Burden of Disease: 2004 Update. Available at https://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/. Accessed September 1, 2018.
- 4.Vigo D, Thornicroft G, Atun R. Estimating the true global burden of mental illness. Lancet Psychiatry. 2016;3:171–178. doi: 10.1016/S2215-0366(15)00505-2. [DOI] [PubMed] [Google Scholar]
- 5.Herman JP, Tasker JG. Paraventricular hypothalamic mechanisms of chronic stress adaptation. Front Endocrinol (Lausanne) 2016;7:137. doi: 10.3389/fendo.2016.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McEwen BS, et al. Mechanisms of stress in the brain. Nat Neurosci. 2015;18:1353–1363. doi: 10.1038/nn.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willner P. The chronic mild stress (CMS) model of depression: History, evaluation and usage. Neurobiol Stress. 2016;6:78–93. doi: 10.1016/j.ynstr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheeta S, Ruigt G, van Proosdij J, Willner P. Changes in sleep architecture following chronic mild stress. Biol Psychiatry. 1997;41:419–427. doi: 10.1016/S0006-3223(96)00058-3. [DOI] [PubMed] [Google Scholar]
- 9.Grønli J, et al. Chronic mild stress affects sucrose intake and sleep in rats. Behav Brain Res. 2004;150:139–147. doi: 10.1016/S0166-4328(03)00252-3. [DOI] [PubMed] [Google Scholar]
- 10.Hegde P, Jayakrishnan HR, Chattarji S, Kutty BM, Laxmi TR. Chronic stress-induced changes in REM sleep on θ oscillations in the rat hippocampus and amygdala. Brain Res. 2011;1382:155–164. doi: 10.1016/j.brainres.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 11.Page GG, Opp MR, Kozachik SL. Sex differences in sleep, anhedonia, and HPA axis activity in a rat model of chronic social defeat. Neurobiol Stress. 2016;3:105–113. doi: 10.1016/j.ynstr.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olini N, et al. Chronic social stress leads to altered sleep homeostasis in mice. Behav Brain Res. 2017;327:167–173. doi: 10.1016/j.bbr.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 13.Ising M, et al. Combined dexamethasone/corticotropin releasing hormone test predicts treatment response in major depression—A potential biomarker? Biol Psychiatry. 2007;62:47–54. doi: 10.1016/j.biopsych.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 14.Baglioni C, et al. Sleep and mental disorders: A meta-analysis of polysomnographic research. Psychol Bull. 2016;142:969–990. doi: 10.1037/bul0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plante DT, et al. Sex-related differences in sleep slow wave activity in major depressive disorder: A high-density EEG investigation. BMC Psychiatry. 2012;12:146. doi: 10.1186/1471-244X-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tesler N, et al. Increased frontal sleep slow wave activity in adolescents with major depression. Neuroimage Clin. 2015;10:250–256. doi: 10.1016/j.nicl.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillai V, Kalmbach DA, Ciesla JA. A meta-analysis of electroencephalographic sleep in depression: Evidence for genetic biomarkers. Biol Psychiatry. 2011;70:912–919. doi: 10.1016/j.biopsych.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Malki K, et al. Pervasive and opposing effects of unpredictable chronic mild stress (UCMS) on hippocampal gene expression in BALB/cJ and C57BL/6J mouse strains. BMC Genomics. 2015;16:262. doi: 10.1186/s12864-015-1431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stankiewicz AM, Goscik J, Majewska A, Swiergiel AH, Juszczak GR. The effect of acute and chronic social stress on the hippocampal transcriptome in mice. PLoS One. 2015;10:e0142195. doi: 10.1371/journal.pone.0142195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surget A, et al. Corticolimbic transcriptome changes are state-dependent and region-specific in a rodent model of depression and of antidepressant reversal. Neuropsychopharmacology. 2009;34:1363–1380. doi: 10.1038/npp.2008.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hervé M, et al. Translational identification of transcriptional signatures of major depression and antidepressant response. Front Mol Neurosci. 2017;10:248. doi: 10.3389/fnmol.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muhie S, et al. Molecular indicators of stress-induced neuroinflammation in a mouse model simulating features of post-traumatic stress disorder. Transl Psychiatry. 2017;7:e1135. doi: 10.1038/tp.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leday GGR, et al. Replicable and coupled changes in innate and adaptive immune gene expression in two case-control studies of blood microarrays in major depressive disorder. Biol Psychiatry. 2018;83:70–80. doi: 10.1016/j.biopsych.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgson K, et al. Transcriptomics and the mechanisms of antidepressant efficacy. Eur Neuropsychopharmacol. 2016;26:105–112. doi: 10.1016/j.euroneuro.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Hepgul N, Cattaneo A, Zunszain PA, Pariante CM. Depression pathogenesis and treatment: What can we learn from blood mRNA expression? BMC Med. 2013;11:28. doi: 10.1186/1741-7015-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill MN, Hellemans KG, Verma P, Gorzalka BB, Weinberg J. Neurobiology of chronic mild stress: Parallels to major depression. Neurosci Biobehav Rev. 2012;36:2085–2117. doi: 10.1016/j.neubiorev.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nollet M, Le Guisquet AM, Belzung C. Models of depression: Unpredictable chronic mild stress in mice. Curr Protoc Pharmacol. 2013;Chapter 5:Unit 5.65. doi: 10.1002/0471141755.ph0565s61. [DOI] [PubMed] [Google Scholar]
- 28.Nollet M, et al. Neurogenesis-independent antidepressant-like effects on behavior and stress axis response of a dual orexin receptor antagonist in a rodent model of depression. Neuropsychopharmacology. 2012;37:2210–2221. doi: 10.1038/npp.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 30.Gilpin AR. Table for conversion of Kendall tau to Spearman rho within the context of measures of magnitude of effect for metaanalysis. Educ Psychol Meas. 1993;53:87–92. [Google Scholar]
- 31.Bjorness TE, Booth V, Poe GR. Hippocampal theta power pressure builds over non-REM sleep and dissipates within REM sleep episodes. Arch Ital Biol. 2018;156:112–126. doi: 10.12871/00039829201833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagot RC, et al. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron. 2016;90:969–983. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreutzmann JC, Havekes R, Abel T, Meerlo P. Sleep deprivation and hippocampal vulnerability: Changes in neuronal plasticity, neurogenesis and cognitive function. Neuroscience. 2015;309:173–190. doi: 10.1016/j.neuroscience.2015.04.053. [DOI] [PubMed] [Google Scholar]
- 34.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160:816–827. doi: 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irwin MR, Opp MR. Sleep health: Reciprocal regulation of sleep and innate immunity. Neuropsychopharmacology. 2017;42:129–155. doi: 10.1038/npp.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu YZ, Wang YX, Jiang CL. Inflammation: The common pathway of stress-related diseases. Front Hum Neurosci. 2017;11:316. doi: 10.3389/fnhum.2017.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malki K, et al. The endogenous and reactive depression subtypes revisited: Integrative animal and human studies implicate multiple distinct molecular mechanisms underlying major depressive disorder. BMC Med. 2014;12:73. doi: 10.1186/1741-7015-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang P, et al. A systems approach identifies networks and genes linking sleep and stress: Implications for neuropsychiatric disorders. Cell Rep. 2015;11:835–848. doi: 10.1016/j.celrep.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anacker C, et al. Glucocorticoid-related molecular signaling pathways regulating hippocampal neurogenesis. Neuropsychopharmacology. 2013;38:872–883. doi: 10.1038/npp.2012.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown MK, Naidoo N. The UPR and the anti-oxidant response: Relevance to sleep and sleep loss. Mol Neurobiol. 2010;42:103–113. doi: 10.1007/s12035-010-8114-8. [DOI] [PubMed] [Google Scholar]
- 41.Brown MK, Strus E, Naidoo N. Reduced sleep during social isolation leads to cellular stress and induction of the unfolded protein response. Sleep. 2017;40:zsx095. doi: 10.1093/sleep/zsx095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Logan RW, et al. Chronic stress induces brain region-specific alterations of molecular rhythms that correlate with depression-like behavior in mice. Biol Psychiatry. 2015;78:249–258. doi: 10.1016/j.biopsych.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClung CA. How might circadian rhythms control mood? Let me count the ways…. Biol Psychiatry. 2013;74:242–249. doi: 10.1016/j.biopsych.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albrecht U. Molecular mechanisms in mood regulation involving the circadian clock. Front Neurol. 2017;8:30. doi: 10.3389/fneur.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldstein AN, Walker MP. The role of sleep in emotional brain function. Annu Rev Clin Psychol. 2014;10:679–708. doi: 10.1146/annurev-clinpsy-032813-153716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pace-Schott EF, Germain A, Milad MR. Sleep and REM sleep disturbance in the pathophysiology of PTSD: The role of extinction memory. Biol Mood Anxiety Disord. 2015;5:3. doi: 10.1186/s13587-015-0018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wassing R, et al. Slow dissolving of emotional distress contributes to hyperarousal. Proc Natl Acad Sci USA. 2016;113:2538–2543. doi: 10.1073/pnas.1522520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldstein-Piekarski AN, Greer SM, Saletin JM, Walker MP. Sleep deprivation impairs the human central and peripheral nervous system discrimination of social threat. J Neurosci. 2015;35:10135–10145. doi: 10.1523/JNEUROSCI.5254-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyce R, Glasgow SD, Williams S, Adamantidis A. Causal evidence for the role of REM sleep theta rhythm in contextual memory consolidation. Science. 2016;352:812–816. doi: 10.1126/science.aad5252. [DOI] [PubMed] [Google Scholar]
- 50.Menz MM, Rihm JS, Büchel C. REM sleep is causal to successful consolidation of dangerous and safety stimuli and reduces return of fear after extinction. J Neurosci. 2016;36:2148–2160. doi: 10.1523/JNEUROSCI.3083-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palagini L, Baglioni C, Ciapparelli A, Gemignani A, Riemann D. REM sleep dysregulation in depression: State of the art. Sleep Med Rev. 2013;17:377–390. doi: 10.1016/j.smrv.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 52.McCarthy A, et al. REM sleep homeostasis in the absence of REM sleep: Effects of antidepressants. Neuropharmacology. 2016;108:415–425. doi: 10.1016/j.neuropharm.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 53.Poland RE, McCracken JT, Lutchmansingh P, Tondo L. Relationship between REM sleep latency and nocturnal cortisol concentrations in depressed patients. J Sleep Res. 1992;1:54–57. doi: 10.1111/j.1365-2869.1992.tb00010.x. [DOI] [PubMed] [Google Scholar]
- 54.Rao U, Hammen CL, Poland RE. Risk markers for depression in adolescents: Sleep and HPA measures. Neuropsychopharmacology. 2009;34:1936–1945. doi: 10.1038/npp.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu WP, Li JD, Colwell CS, Zhou QY. Decreased REM sleep and altered circadian sleep regulation in mice lacking vasoactive intestinal polypeptide. Sleep. 2011;34:49–56. doi: 10.1093/sleep/34.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Born J, Kellner C, Uthgenannt D, Kern W, Fehm HL. Vasopressin regulates human sleep by reducing rapid-eye-movement sleep. Am J Physiol. 1992;262:E295–E300. doi: 10.1152/ajpendo.1992.262.3.E295. [DOI] [PubMed] [Google Scholar]
- 57.Narwade SC, Mallick BN, Deobagkar DD. Transcriptome analysis reveals altered expression of memory and neurotransmission associated genes in the REM sleep deprived rat brain. Front Mol Neurosci. 2017;10:67. doi: 10.3389/fnmol.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luppi PH, Peyron C, Fort P. Role of MCH neurons in paradoxical (REM) sleep control. Sleep. 2013;36:1775–1776. doi: 10.5665/sleep.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loh DH, Abad C, Colwell CS, Waschek JA. Vasoactive intestinal peptide is critical for circadian regulation of glucocorticoids. Neuroendocrinology. 2008;88:246–255. doi: 10.1159/000140676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goncharova ND. Stress responsiveness of the hypothalamic-pituitary-adrenal axis: Age-related features of the vasopressinergic regulation. Front Endocrinol (Lausanne) 2013;4:26. doi: 10.3389/fendo.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buysse DJ, et al. Sleep and treatment response in depression: New findings using power spectral analysis. Psychiatry Res. 2001;103:51–67. doi: 10.1016/s0165-1781(01)00270-0. [DOI] [PubMed] [Google Scholar]
- 62.Le TT, et al. Identification and replication of RNA-seq gene network modules associated with depression severity. Transl Psychiatry. 2018;8:180. doi: 10.1038/s41398-018-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woo HI, Lim SW, Myung W, Kim DK, Lee SY. Differentially expressed genes related to major depressive disorder and antidepressant response: Genome-wide gene expression analysis. Exp Mol Med. 2018;50:92. doi: 10.1038/s12276-018-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao L, et al. rTMS ameliorated depressive-like behaviors by restoring HPA axis balance and prohibiting hippocampal neuron apoptosis in a rat model of depression. Psychiatry Res. 2018;269:126–133. doi: 10.1016/j.psychres.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 65.Skorucak J, Arbon EL, Dijk DJ, Achermann P. Response to chronic sleep restriction, extension, and subsequent total sleep deprivation in humans: Adaptation or preserved sleep homeostasis? Sleep. 2018;41:zsy078. doi: 10.1093/sleep/zsy078. [DOI] [PubMed] [Google Scholar]
- 66.Laing EE, Möller-Levet CS, Dijk DJ, Archer SN. Identifying and validating blood mRNA biomarkers for acute and chronic insufficient sleep in humans: A machine learning approach. Sleep. September 24, 2018 doi: 10.1093/sleep/zsy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Husse J, et al. Tissue-specific dissociation of diurnal transcriptome rhythms during sleep restriction in mice. Sleep. 2017;40:zsx068. doi: 10.1093/sleep/zsx068. [DOI] [PubMed] [Google Scholar]
- 68.Puthalakath H, et al. Bmf: A proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293:1829–1832. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- 69.Allen J, Romay-Tallon R, Brymer KJ, Caruncho HJ, Kalynchuk LE. Mitochondria and mood: Mitochondrial dysfunction as a key player in the manifestation of depression. Front Neurosci. 2018;12:386. doi: 10.3389/fnins.2018.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meerlo P, Mistlberger RE, Jacobs BL, Heller HC, McGinty D. New neurons in the adult brain: The role of sleep and consequences of sleep loss. Sleep Med Rev. 2009;13:187–194. doi: 10.1016/j.smrv.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Navarro-Sanchis C, Brock O, Winsky-Sommerer R, Thuret S. Modulation of adult hippocampal neurogenesis by sleep: Impact on mental health. Front Neural Circuits. 2017;11:74. doi: 10.3389/fncir.2017.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fitzsimons CP, et al. Circadian and ultradian glucocorticoid rhythmicity: Implications for the effects of glucocorticoids on neural stem cells and adult hippocampal neurogenesis. Front Neuroendocrinol. 2016;41:44–58. doi: 10.1016/j.yfrne.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 73.Lucassen PJ, et al. Regulation of adult neurogenesis and plasticity by (early) stress, glucocorticoids, and inflammation. Cold Spring Harb Perspect Biol. 2015;7:a021303. doi: 10.1101/cshperspect.a021303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Labonté B, et al. Sex-specific transcriptional signatures in human depression. Nat Med. 2017;23:1102–1111. doi: 10.1038/nm.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma XC, et al. Long-lasting antidepressant action of ketamine, but not glycogen synthase kinase-3 inhibitor SB216763, in the chronic mild stress model of mice. PLoS One. 2013;8:e56053. doi: 10.1371/journal.pone.0056053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gray JD, Rubin TG, Hunter RG, McEwen BS. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2014;19:1171–1178. doi: 10.1038/mp.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khan AR, Hansen B, Wiborg O, Kroenke CD, Jespersen SN. Diffusion MRI and MR spectroscopy reveal microstructural and metabolic brain alterations in chronic mild stress exposed rats: A CMS recovery study. Neuroimage. 2018;167:342–353. doi: 10.1016/j.neuroimage.2017.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin M, Hou G, Zhao Y, Yuan TF. Recovery of chronic stress-triggered changes of hippocampal glutamatergic transmission. Neural Plast. 2018;2018:9360203. doi: 10.1155/2018/9360203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hasan S, van der Veen DR, Winsky-Sommerer R, Dijk DJ, Archer SN. Altered sleep and behavioral activity phenotypes in PER3-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2011;301:R1821–R1830. doi: 10.1152/ajpregu.00260.2011. [DOI] [PubMed] [Google Scholar]
- 80.Petit-Demouliere B, Chenu F, Bourin M. Forced swimming test in mice: A review of antidepressant activity. Psychopharmacology (Berl) 2005;177:245–255. doi: 10.1007/s00213-004-2048-7. [DOI] [PubMed] [Google Scholar]
- 81.Moy SS, et al. Sociability and preference for social novelty in five inbred strains: An approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 82.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 83.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 84.Maher JM, Markey JC, Ebert-May D. The other half of the story: Effect size analysis in quantitative research. CBE Life Sci Educ. 2013;12:345–351. doi: 10.1187/cbe.13-04-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.