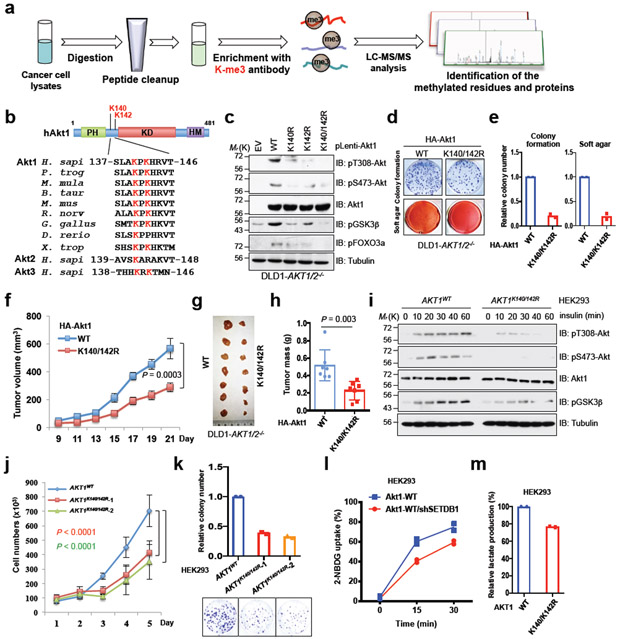
Fig. 1. Akt methylation promotes its activity and oncogenic functions.
a, A schematic workflow of IAP-LC-mass spectrometry (MS)/MS experiments. OVCAR5 cell lysates were proteolytically digested to perform IAP-LC-MS/MS assays. b, Alignment of MS-characterized Akt1 putative methylation residues among different species, Akt2 and Akt3. c, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from DLD1-AKT1/2−/− cells infected with indicated Akt1 encoding virus and selected with hygromycin (200 μg/ml) for 72 hrs before harvesting. Data shown represent two independent experiments. d-e, cells generated in c were subjected to colony formation and soft agar assays. The experiment was performed twice independently with three repeats, and exhibited similar results (d). Representative images were shown in d and relative colony numbers derived from two independent experiments were plotted in e. f-h, Cells generated in (c) were subjected to mouse xenograft assays. Tumor sizes were monitored (f), and dissected tumors were weighed (g, h). Error bars in f and h are mean ± s.e.m, n = 7 mice. i-k, K140R and K142R mutations of Akt1 were genetically engineered in HEK293 cells by the CRISPR/CAS9-based technique. Resulting cells were serum-starved for 12 hrs, and harvested for IB analysis at different time points after stimulation with insulin (100 nM) (i). The experiment was performed twice independently with similar results (i). Cells generated in (i) were assessed for proliferation (j) assays. The experiment in j was performed three times independently and exhibited similar results. Error bars in j are mean ± s.e.m, n = 3 independent experiments. k,l,m, Cells generated in i were subjected to colony formation (k), glucose uptake (l) and lactate production (m) assays. The experiment was performed twice independently with three repeats, and exhibited similar results (k,l,m). Relative colony numbers, glucose and lactate levels derived from two independent experiments were plotted in (k,l,m). Two-way ANOVA analysis was performed in (f,j) to calculate the P value. Detailed statistical tests are described in the Methods. Source data for e, f, h and j-m are shown in Supplementary Table 2. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.