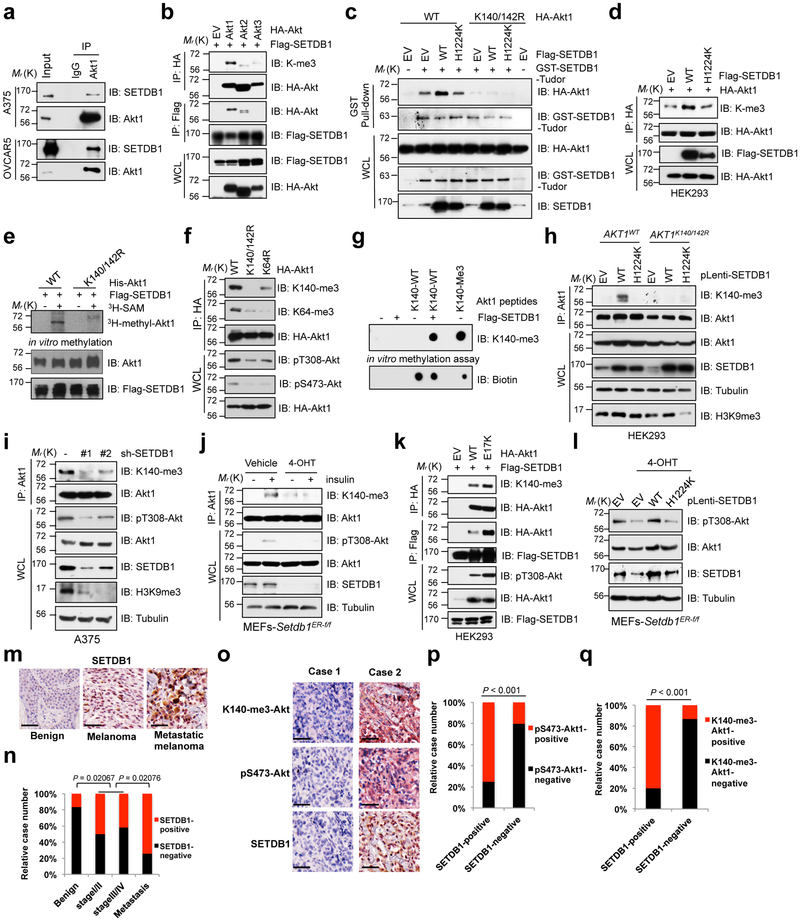
Fig. 3. SETDB1 methylates Akt on K140 and K142 to promote its kinase activity.
a-d,f,h, IB analysis of immunoprecipitates (IP), GST pulldown products and WCL derived from A375 and OVCAR5 (a) or HEK293 cells transfected with indicated constructs (b-d, f) or AKT1K140/142R-KI cells stably expressed WT or catalytically inactive SETDB1-H1224K (d). IgG was used as a negative control. e, In vitro methylation assays were performed with recombinant His-Akt1 proteins purified from insect cells as substrates and purified Flag-SETDB1 from HEK293T cells as the source of methyltransferase in the presence/absence of 3H-SAM. g, In vitro methylation assays were performed with IP Flag-SETDB1 derived from HEK293T cells as the source of methyltransferase, and the synthetic Akt1 peptides containing K140 and K142 as the substrate. Akt1-K140-me3 peptides were used as a positive control. h, IB analysis of IP products and WCL derived from A375 cells lentivirally transfected with shRNAs against SETDB1. Resulting cells were selected with puromycin for 72 hrs before harvesting. j, Setdb1 conditional knockout MEFs were treated with or without 4-OHT (500 nM) for 48 hrs to deplete endogenous Setdb1, then resulting cells were serum-starved for another 20 hrs and stimulated with insulin (100 nM) for 15 min before being harvested and subjected to IP and IB analysis. k, IB analysis of IP products and WCL derived from HEK293 cells transfected with indicated constructs. l, Setdb1 conditional knockout MEFs were infected with WT or H1224K-SETDB1 encoding virus and selected with puromycin for 72 hrs, then treated with or without 4-OHT (500 nM) for another 48 hrs before harvesting for IB analysis. m-q, Image illustration of the immunohistochemistry (IHC) staining for SETDB1, pS473-Akt1 and K140-me3-Akt1 in melanoma TMA (m,o). Scale bar, 50 μm. The distribution of SETDB1 staining was plotted in (n) (n = 97 tissue specimens). The correlation of pS473-Akt or K140-me3-Akt with SETDB1 were plotted in (p,q) (n = 95 tissue specimens). Western-blots in (a-l) were performed twice, independently, with similar results. P values were calculated using Chi-Square test in (n-q). Statistical source data for n-q are shown in Supplementary Table 2. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.