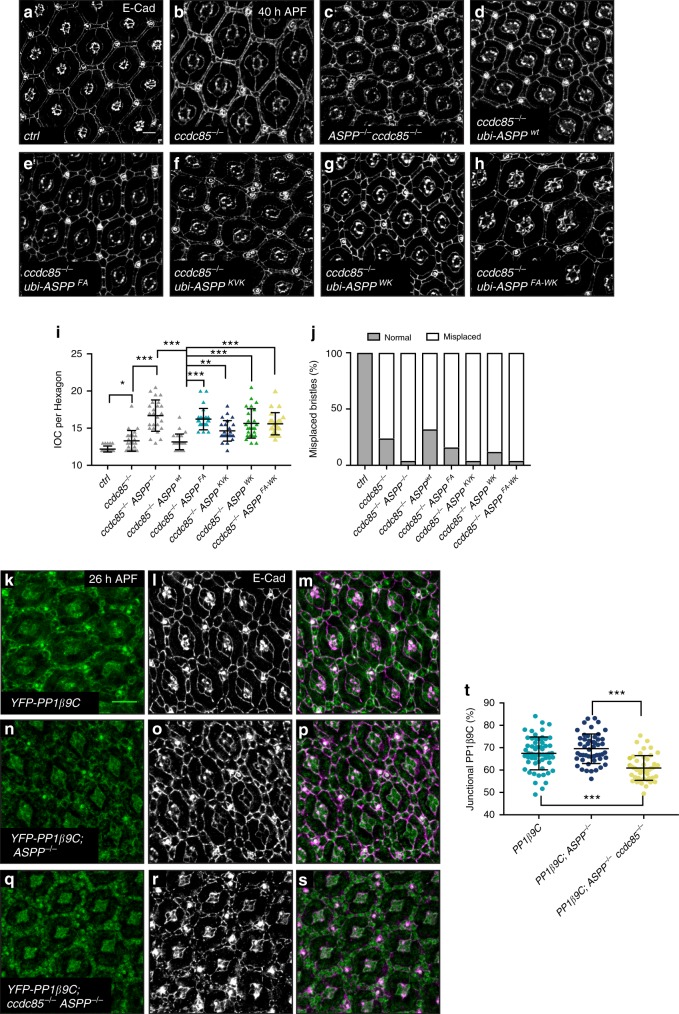
Fig. 9.
The ASPP complex promotes PP1β9C and PP1α96A AJ localization. a–h Confocal X–Y sections of pupal retinas at 40 h APF stained with anti-E-cad antibodies to mark cell outlines. Scale bar = 10 µm. a, b ccdc85 (b) mutant flies have defects in eye development compared to wild-type flies (a) (13.30 ± 1.40 vs 12.2 ± 0.4). c Mutation of ASPP in ccdc85 mutant flies worsens the phenotype (16.7 ± 2.10). d–g Expression of ASPP under the ubiquitin 63E (ubi) promoter in ASPP, ccdc85 mutants rescues the phenotype (d), but overexpression of ASPPFA (e), ASPPKVK (f), ASPP WK (g) or ASPPFA-WK (h) do not (13.16 ± 1.058 vs 16.22 ± 1.44, 14.64 ± 1.38, 15.64 ± 1.98 and 15.60 ± 1.49) (see Supplementary table 1 for full genotypes). i Quantification of inter-ommatidial cells (IOCs) in retinas of the indicated genotypes at 40 h after puparium formation (APF). Error bars represent the standard deviation. A one-way ANOVA test was carried out to determine if the differences among means were significantly different from each other. p-values were adjusted using a Bonferroni correction. Significant differences are marked. *** indicates p < 0.0001, ** indicates p < 0.01 and * indicates p < 0.05 (n = 25 hexagons from four retinas). j Quantification of bristle misplacement. (n = 25 from four retinas). k–s Confocal X–Y sections of pupal retinas at 26 h APF showing endogenous YFP-tagged PP1β9C and stained with E-Cad. Scale bar = 10 µm. k–m PP1β9C localises at the cell–cell junctions (67.31% ± 7.45). n–p ASPP loss does not change the localisation of PP1-β9C (69.23% ± 6.29). q–s Loss of ccdc85 in an ASPP mutant background mislocalises PP1β9C from the cell–cell junctions (60.92% ± 5.51). t Quantification of percentage of junctional PP1β9C of the indicated genotypes at 26 h APF. Error bars represent the standard deviation. For quantifications of PP1β9C intensity, two regions of interest were drawn surrounding the junctions of a cell (ROITOTAL and ROICYTOPLASM). Junctional fraction was quantified by (ROITOTAL – ROICYTOPLASM). Values were normalised to 100% for PP1β9C. One-way ANOVA test was carried out to determine if the differences among means were significantly different from each other. p-values were adjusted using a Bonferroni correction. Significant differences are marked. *** indicates p < 0.0001 (n = 50 cells from four retinas)