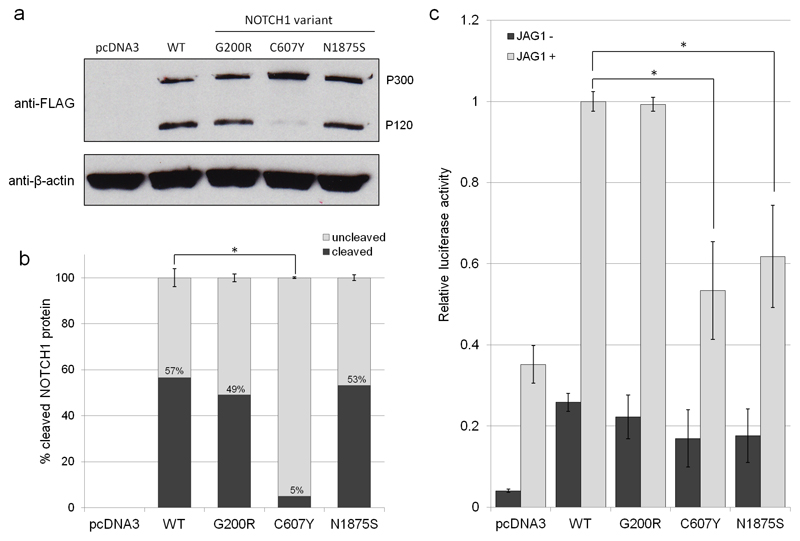
Figure 3.
(a) Immunoblot for FLAG to determine the expression and S1 cleavage of NOTCH1 variants p.G200R, p.C607Y and p.N1875S in comparison to WT NOTCH1 following overexpression in HeLa cells. The two bands at 300 kDa (P300) and 120 kDa (P120) represent the full length and the S1-cleaved NOTCH1 protein. β-actin was used as a loading control. (b) Quantification of the percentage of S1 cleaved versus uncleaved NOTCH1 protein for WT NOTCH1 and NOTCH variants p.G200R, p.C607Y and p.N1875S. Error bars: mean ±SEM from three biological replicates and statistical significance was determined using two-tailed paired t-tests. (c) The effect of rare, deleterious NOTCH1 variants on Jagged-induced NOTCH signalling levels. NOTCH signalling activity was measured using a luciferase-based reporter system (RBPJ). HeLa cells were cultured with or without immobilised JAG1 ligand and co-transfected with RBPJ reporter constructs and WT NOTCH1, p.G200R, p.C607Y or p.N1875S. Firefly luciferase readings were normalised to Renilla luciferase readings to control for transfection efficiency and cell number. RBPJ activity was expressed relative to WT NOTCH1 for comparison. Error bars: mean ±SEM from four biological replicates, each with three technical replicates. Statistical significance was assessed using two-tailed paired t-tests and the Hochberg step-up procedure to control for family-wise error rate.