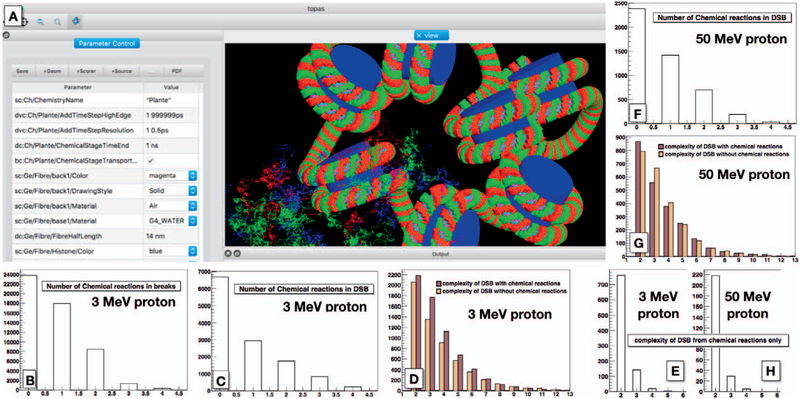
FIG. 10.
Simulation of a chromatid fiber irradiated with 3 and 50 MeV protons, including the chemistry of radiolysis. The center shows a representation of the simulated geometry and particle tracks, including the propagation of chemical species. Panels B–E and F–H: The results for 3 and 50 MeV proton irradiations, respectively. The plots demonstrate the effects of including chemical reactions. Here, only interactions with hydroxyl radicals are scored. Approximately one half of the DSBs include a reaction with hydroxyl, which not only influences the overall number but also the complexity of DNA damage (defined as number of lesions within a DSB).