Figure 2.
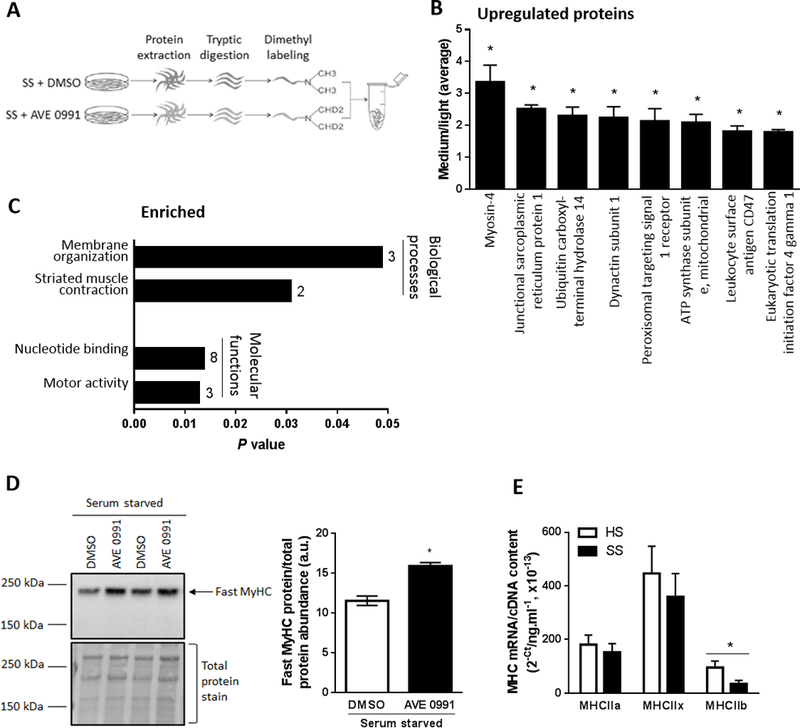
Proteomic profiling to identify proteins and pathways regulated by AVE0991 treatment of serum starved C2C12 myotubes. (A) Schematic of the experiment, with proteins from serum starved (SS) C2C12 myotubes treated with vehicle control (DMSO]) or AVE0991 ]extracted using lysis buffer containing 1% Triton-X 100, tryptic digested and purified using SPE cartridges. Peptides obtained from DMSO and AVE0991 treated serum starved myotubes were labeled with normal (light) or deuterated (medium) formaldehyde, respectively. The labelled peptides were mixed together and analysed by LC-MS/MS using an LTQ Orbitrap Elite mass spectrometer. (B) Upregulated proteins and (C) enriched biological processes and molecular functions representing upregulated proteins obtained from DAVID database in AVE0991 treated serum starved myotubes. Numbers next to solid bars in C indicates counts of proteins annotated to the indicated processes and functions (unpaired t test; *P<0.05 Serum starved+AVE0991 vs. Serum starved+DMSO; n=3). (D) Representative western blot and corresponding quantification of fast myosin heavy chain (MyHC) confirming upregulation of myosin-4, the top ranked upregulated protein, in AVE0991 treated serum starved myotubes (unpaired t test; *P<0.05; n=6). (E) Gene expression of the fast myosin heavy chain isoforms MHCIIa, MHCIIx and MHCIIb in C2C12 myotubes incubated for 48 h in HS or serum-free media (SS, unpaired t test; *P<0.05; n=6).
