Fig. 3.
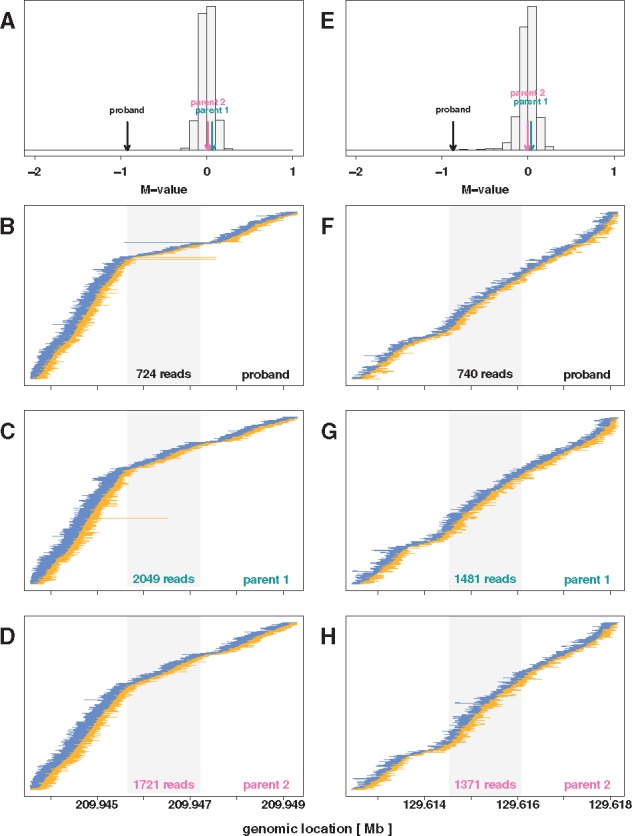
Data underlying inferred de novo heterozygous deletions in two probands. [Left Column] Evidence for a de novo heterozygous deletion on chromosome 1 for the proband in family DS10826. (A) The average of the M scores of the proband (-0.93, black arrow), the parents (0.06 and 0.01, green and pink arrows, respectively) and all other subjects (gray histogram) between loci 209 945 655 and 209 947 210 on chromosome 1. The proband’s average of the M scores near −1, compared to the values near zero for all other samples including the parents, is consistent with a de novo deletion of one allele in this region. (B–D) Read-pairs observed among the members of family DS10826 near the region with the de novo heterozygous deletion. The read-pair locations, mapped to the hg19 reference genome, are shown as thick ends connected by thin lines (positive strands shown in yellow, negative strands shown in blue), and sorted by beginning location of mate 1 of the read-pair (e.g. yellow lines are left aligned, blue lines are right aligned). Read-pairs mapped far apart, apparent as a long line, are indicative of a deletion between the ends. A Z-shaped signature of read pairs flanked by such discordant reads as seen for the proband is strong evidence for a 1-copy DNA deletion. The gray region in these panels indicate the inferred 1556 bp heterozygous de novo deletion region in the proband’s genome. The number at the bottom of the grey regions in each panel indicates the total number of reads mapped to the inferred de novo deletion. [Right Column] A possible false positive identification of a de novo heterozygous deletion on chromosome 8 for the proband in family DS12329. (E) The average of the M scores of the proband (−0.87), the parents (0.035 and −0.007, green and pink arrows, respectively), and all other subjects (gray histogram) between loci 129 614 522 and 129 616 078 on chromosome 8. The proband’s average of the M scores near −1, compared to the values near zero for all other samples including the parents, is consistent with a de novo deletion of one allele in this region. (F–H) Read-pairs observed among the members of family DS12329 near the region with the inferred de novo heterozygous deletion. The absence of discordant reads and the Z-shaped signature is evidence against a 1-copy DNA deletion (Color version of this figure is available at Bioinformatics online.)
