Abstract
Field studies have shown that plants growing next to herbivore-infested plants acquire higher resistance to herbivore damage. This increased resistance is partly due to regulation of plant gene expression by volatile organic compounds (VOCs) released by plants that sense environmental challenges such as herbivores. The molecular basis for VOC sensing in plants, however, is poorly understood. Here, we report the identification of TOPLESS-like proteins (TPLs) that have VOC-binding activity and are involved in VOC sensing in tobacco. While screening for volatiles that induce stress-responsive gene expression in tobacco BY-2 cells and tobacco plants, we found that some sesquiterpenes induce the expression of stress-responsive genes. These results provided evidence that plants sense these VOCs and motivated us to analyze the mechanisms underlying volatile sensing using tobacco as a model system. Using a pulldown assay with caryophyllene derivative–linked beads, we identified TPLs as transcriptional co-repressors that bind volatile caryophyllene analogs. Overexpression of TPLs in cultured BY-2 cells or tobacco leaves reduced caryophyllene-induced gene expression, indicating that TPLs are involved in the responses to caryophyllene analogs in tobacco. We propose that unlike animals, which use membrane receptors for sensing odorants, a transcriptional co-repressor plays a role in sensing and mediating VOC signals in plant cells.
Keywords: receptor, plant biochemistry, ligand-binding protein, transcription regulation, transcription corepressor, odorant, chemical sensing, plant volatile perception, volatile organic compounds (VOCs), defense induction, transcriptional co-repressor protein, transcription factor, gene regulation, olfaction
Introduction
For terrestrial animals, odorants or volatile organic compounds (VOCs)5 possess important biological and ecological information such as food, predator, and species. Sensing these chemical cues, animals take appropriate behavior such as attraction or avoidance, ensuring their survival. Plants also have to acquire information from the external environment and take appropriate action for survival. Defense or stress-related genes are up-regulated upon exposure to specific VOCs to prepare for environmental change in plants (1–4). For example, defense genes are induced in healthy lima bean leaves upon exposure to VOCs from infested leaves, but not from healthy or artificially wounded leaves (5). In addition, VOCs released from infested leaves prime neighboring plants for direct and indirect defense against future herbivore attack (6). VOCs are also used as cues for host selection and location by parasitic plants (7). Several individual compounds from host plants also show attractiveness to parasitic plants. Regardless of accumulated evidence for the VOC effects in plants, a molecular basis for the VOC detection by plant cells has not been revealed.
In animals, VOCs are recognized by odorant receptors in the olfactory neural system that constitutes the largest G protein–coupled receptor (GPCR) family. In contrast, plants have only a few GPCR genes that appear to have different functions (8). It has been unclear how VOCs are sensed and the information is converted to signals that induce the specific responses in plant cells at the level of receptors. It is possible that plants have acquired a different strategy to detect external VOCs because they cannot move. This hypothesis is supported by the fact that Dictyostelium, along the evolutionary branch soon after the plant-animal split, has a large GPCR family that possibly mediates various behaviors such as chemotaxis (9). The aim of this study is to explore plant VOC-binding proteins that detect external VOCs and are involved in regulation of gene expression.
Results
Volatile-induced gene expression in BY-2 cell
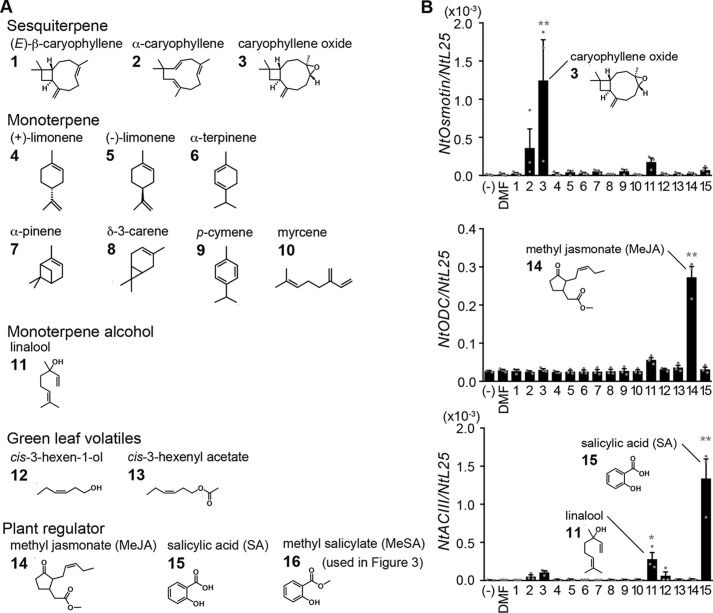
We first examined whether VOCs released from plants induce gene expression in tobacco BY-2 cell cultures (10). The tested VOCs included three sesquiterpenes, seven monoterpenes, a monoterpene alcohol, and two green leaf volatiles (Fig. 1A) that are released from damaged plants (11, 12) and thus are expected to induce defense responses. Methyl jasmonate (MeJA) and salicylic acid (SA) were used as controls for gene expression because these are known as plant regulators. VOCs were added at a final concentration of 1 mm to BY-2 cell culture, and after 3 h, cells were collected for RNA extraction. The expression of three genes was quantitated by quantitative PCR (Q-PCR). Expression of Osmotin, one of the pathogenesis-related proteins involved in infection resistance (13), was induced significantly by caryophyllene oxide and weakly by α-caryophyllene in BY-2 cells (Fig. 1B). Ornithine decarboxylase (ODC), a jasmonic acid (JA)-related gene, was induced by MeJA as described previously (14) but not by the tested VOCs. Acidic chitinase III (ACIII), a SA-related gene, was induced by SA, consistent with previous studies (15), weakly by linalool, but not by the other VOCs tested. These results demonstrate high specificity and selectivity in gene induction by VOCs, suggesting the presence of specific receptors for VOCs that link to individual regulation of each gene.
Figure 1.
Gene expression profiles induced by various VOCs in BY-2 cells. A, chemical structures of VOCs and plant regulators used for structure–activity relationships in B and Fig. 3B. The VOCs were classified into five groups. Numbers placed at the top left-hand corner of each structure correspond to labels in B and Fig. 3B. B, structure–activity relationships of VOC-induced gene expression in BY-2 cells. BY-2 cells were exposed to DMF (0.1%), MeJA (0.1 mm), SA (0.1 mm), or other VOCs (1 mm; see A) for 3 h. Top, NtOsmotin; middle panel, NtODC (jasmonic acid-related gene); bottom, NtACIII (salicylic acid-related gene). Expression of each gene was normalized to NtL25. Error bars, S.E. (n = 3). Statistics were calculated using one-way ANOVA and Dunnett's post hoc test: *, p < 0.05; **, p < 0.01.
Time course and dose-dependent responses of BY-2 cells to caryophyllene structural analogues
We looked into NtOsmotin induction by caryophyllene oxide and its structural analogs in more detail because caryophyllene oxide, (E)-β-caryophyllene, and α-caryophyllene are produced and emitted when tobacco is eaten by insects (12), and (E)-β-caryophyllene is also released from roots to attract nematodes that infect herbivore larvae when maize roots are damaged by larvae (16, 17). First, we examined time course of NtOsmotin expression induced by caryophyllene structural analogues. (E)-β-caryophyllene, α-caryophyllene, or caryophyllene oxide (1 mm, final concentration) was incubated with BY-2 cells up to 12 h. BY-2 cells were collected for RNA extraction at each time point, and Q-PCR analysis was used to investigate the expression of three genes. Maximum induction of NtOsmotin by caryophyllene structural analogues was at 3–6 h of incubation in BY-2 cells (Fig. 2A).
Figure 2.
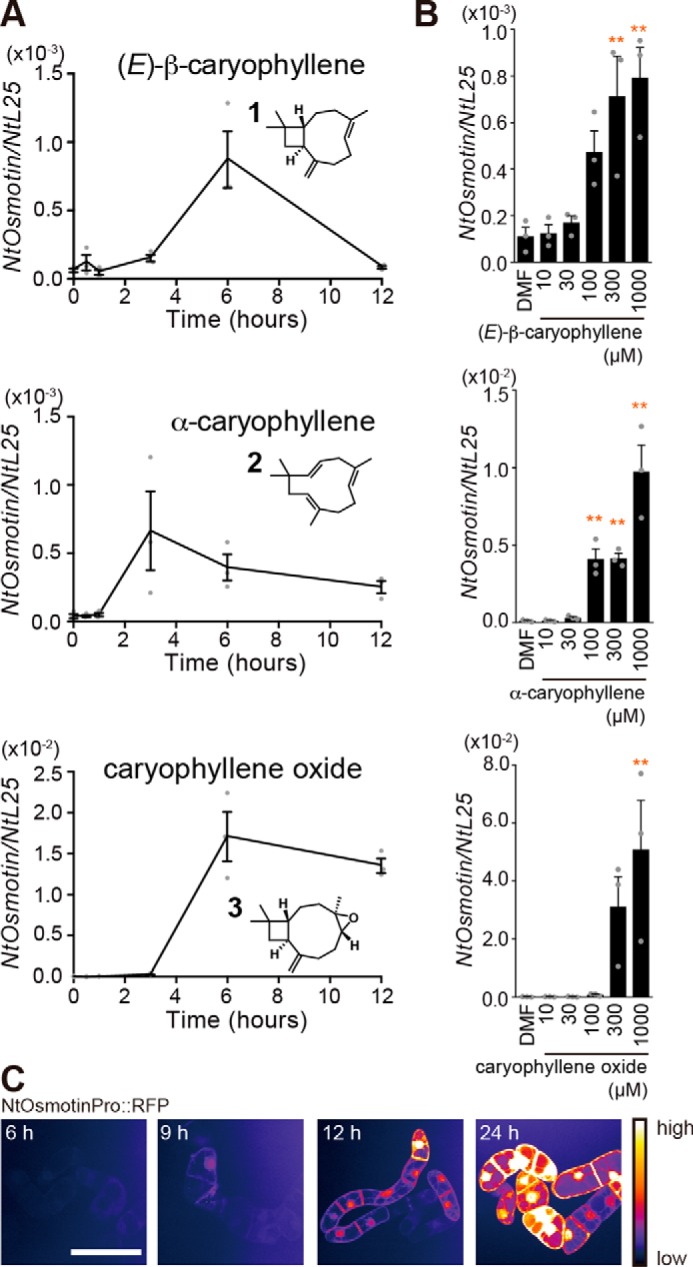
Gene expression profiles induced by caryophyllene structural analogues in BY-2 cells. A, time course of NtOsmotin expression in BY-2 cells treated with caryophyllene structural analogues. BY-2 cells were exposed to analogues (1 mm each) up to 12 h. Error bars, S.E. (n = 3). B, dose-dependent gene expression induced by caryophyllene structural analogues. BY-2 cells were exposed to DMF (0.1%) or caryophyllene structural analogues (10–1000 μm) for 6 h. Top, (E)-β-caryophyllene; middle, α-caryophyllene; bottom, caryophyllene oxide. Error bars, S.E. (n = 3). Statistics were calculated using one-way ANOVA and Dunnett's post hoc test: *, p < 0.05; **, p < 0.01. C, reporter gene assay of NtOsmotin expression using transgenic BY-2 cell line. RFP signals induced by caryophyllene oxide (1 mm) are shown as pseudocolored images (white, highest expression). Scale bar, 50 μm.
Next, we measured sensitivity of BY-2 cells to caryophyllene structural analogues. The responses were dose-dependent with a threshold concentration of a few hundred micromolar (Fig. 2B). To visualize the gene induction, we generated transgenic BY-2 cell lines expressing RFP under the control of the NtOsmotin promoter and performed a reporter gene assay. The RFP signal was first observed at 9 h of incubation with caryophyllene oxide and continuously increased (Fig. 2C). All of these results suggest that BY-2 cells can be used as a model system to investigate a mechanism of VOC detection in plants, especially for caryophyllene structural analogues.
Responses of tobacco plants to caryophyllene structural analogues
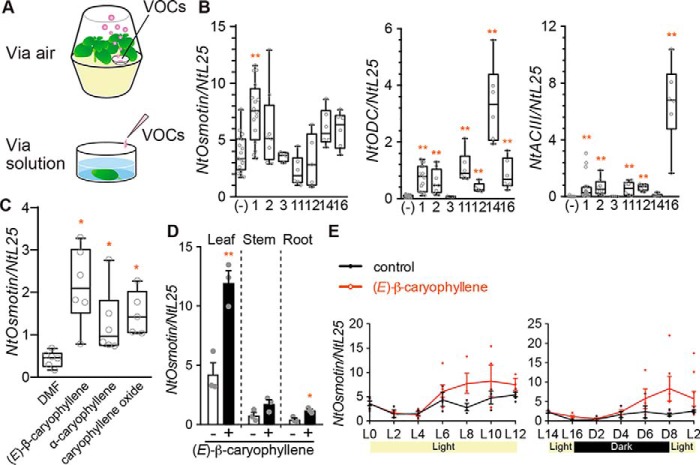
To examine whether caryophyllene structural analogues induce NtOsmotin expression in planta, we treated 4-week-old tobacco plants grown on Murashige and Skoog medium in pots with selected VOCs (Fig. 3A). After 8 h of exposure to VOCs, leaves were harvested for RNA extraction and then analyzed for gene expression via Q-PCR. Among the tested VOCs, (E)-β-caryophyllene induced the highest expression of NtOsmotin in tobacco plants (Fig. 3B). NtODC and NtACIII genes were most induced by MeJA and methyl salicilate, respectively (Fig. 3B). These results were fairly consistent with those of BY-2 cells, although the results in planta were more variable. The response specificity in NtOsmotin induction was slightly different; the order of activity in BY-2 cells was caryophyllene oxide > α-caryophyllene > (E)-β-caryophyllene, whereas that in planta was (E)-β-caryophyllene > α-caryophyllene > caryophyllene oxide. Also, α-caryophyllene and (E)-β-caryophyllene induced NtODC and NtACIII genes significantly in planta but did not in BY-2. We quantified the amounts of caryophyllene structural analogues that BY-2 cells took up from a liquid medium by GC/MS analysis. Caryophyllene oxide was taken up by BY-2 cells better than (E)-β-caryophyllene and α-caryophyllene (see Fig. S1, A and B). In contrast, the amount of caryophyllene in the headspace of an enclosed plant pot was much larger than that of caryophyllene oxide, suggesting that caryophyllene was much more volatile than caryophyllene oxide (see Fig. S1, A and C). Thus, the difference in the response specificity between BY-2 cells and plants turned out to be due to differences in volatility and cellular uptake. Indeed, when tobacco leaves were immersed in VOC solutions, all three caryophyllene analogs induced NtOsmotin (Fig. 3C). Together, our results demonstrated that (E)-β-caryophyllene is sensed by tobacco plants, consistent with previous studies showing that (E)-β-caryophyllene is used as a signal in plants (16, 17). Among leaves, stems, and roots, the highest induction of NtOsmotin was observed in leaves (Fig. 3D). (E)-β-caryophyllene induced NtOsmotin expression regardless of light conditions (Fig. 3E).
Figure 3.
Gene expression profiles induced by various VOCs in tobacco plants. A, experimental setup. Top, the method for B and Fig. 5F. Bottom, the method for C. B, structure–activity relationships of VOC-induced gene expression in tobacco plants. Plants were exposed to each compound (40 μl; see compound numbers in Fig. 1A) for 8 h. Left, NtOsmotin; middle, NtODC (jasmonic acid-related gene); right, NtACIII (salicylic acid–related gene). Expression of each gene was normalized to NtL25 (n = 4–17). Whiskers indicate 1.5 times the interquartile range. Statistics were calculated using the Kruskal–Wallis test and post hoc test (Steel test): *, p < 0.05; **, p < 0.01. C, NtOsmotin expression induced by immersing leaves in solutions of caryophyllene structural analogues (1 mm) or DMF (0.1%) for 8 h (n = 5–6). Whiskers indicate 1.5 times the interquartile range. Statistics were calculated using the Kruskal–Wallis test and post hoc test (Steel test): *, p < 0.05. D, NtOsmotin expression induced by (E)-β-caryophyllene in different organs. White bars, control group; black bars, experimental group. Error bars, S.E. (n = 3). Statistics were calculated using unpaired Student's t test: *, p < 0.05; **, p < 0.01. E, time course of NtOsmotin expression induced by (E)-β-caryophyllene under different light conditions. Left, from light phase 0-h time point (L0) to L12; right, L14–L2. Error bars, S.E. (n = 3–9).
Identification of TOPLESS-like proteins as a binding protein for caryophyllene structural analogues in tobacco cells
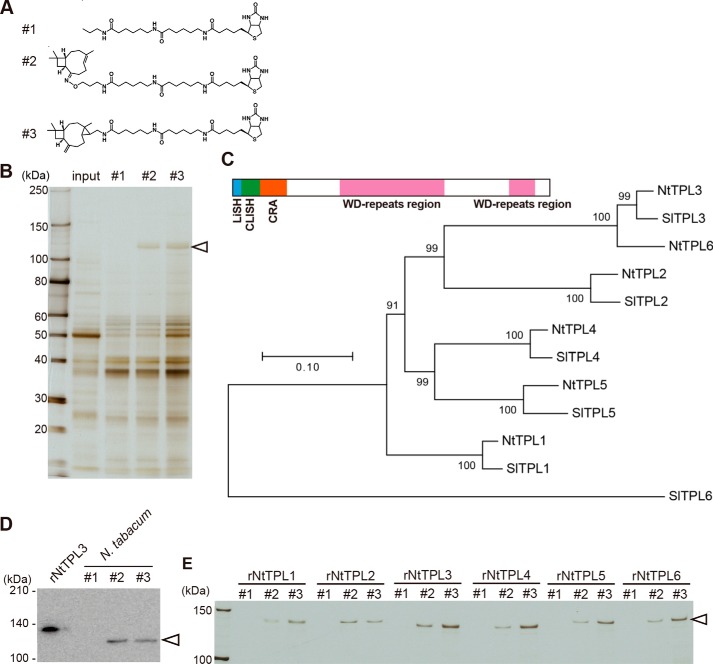
We next attempted to identify receptor-like molecules that recognized the molecular structure of caryophyllene analogues and sent a signal to the nucleus leading to NtOsmotin induction. To fish out a binding protein, we prepared caryophyllene-linked beads for a pulldown assay. Because we did not know which part of the caryophyllene structure is recognized, two types of biotinylated caryophyllene derivatives were synthesized and immobilized onto streptavidin-coated beads (Fig. 4A). When extracts of tobacco leaves were applied to the beads, a 120-kDa protein was specifically pulled down (Fig. 4B). In-gel digestion and LC-MS/MS analysis suggested that the 120-kDa protein was similar to Solanum lycopersicum TOPLESS (TPL)-like protein 3 (SlTPL3). Nicotiana benthamiana TPL genes were then identified in the draft genome sequence using all SlTPLs as queries. Finally, in Nicotiana tabacum, 5′ and 3′ ends of TPL-like genes were PCR-amplified, and six TPL-like genes were sequenced. We named the six NtTPLs as NtTPL1 through -6 based on a phylogenetic tree (Fig. 4C; see Fig. S2). To confirm that NtTPLs indeed bind caryophyllene, we raised an antibody against NtTPL3 and investigated whether NtTPLs were detected in the pulled down samples by Western blotting. The 120-kDa protein was clearly stained by anti-NtTPL3 antibody (Fig. 4D). In addition, GST fusion proteins for all NtTPLs bound the caryophyllene derivative–linked beads (Fig. 4E). These results demonstrate that NtTPLs are binding proteins for caryophyllene structural analogues in tobacco leaves.
Figure 4.
Binding of TOPLESS to caryophyllene-linked beads. A, structure of biotinylated probes. #1, propylamine (control); #2, (E)-β-caryophyllene oxime derivative; #3, (E)-β-caryophyllene derivative with three-membered ring. B, pulldown assay using (E)-β-caryophyllene-linked beads. Total protein extracts (1 mg of protein) of N. tabacum leaves were incubated with caryophyllene-linked beads (10 μl) (shown in A) for 1 h at 4 °C. A 120-kDa protein (arrowhead) specifically bound to beads 2 and 3 but not 1. C, phylogenetic tree of TPL-related proteins in N. tabacum and S. lycopersicum. The structure of the NtTPL3 protein is shown above the tree. NtTPLs are predicted by PROSITE to have a LisH domain (blue), CLISH domain (green), CRA domain (orange), and WD40 repeats (light red). D, Western blots of extracts from N. tabacum incubated with probes shown in A. The leftmost lane shows recombinant GST-NtTPL3 protein (rNtTPL3). The 120-kDa protein of N. tabacum extracts that bound to probes 2 and 3 (arrowhead) was immunostained with anti-NtTPL3 antibody. E, recombinant NtTPLs that bound to caryophyllene-linked beads 2 and 3. Six recombinant GST-NtTPLs (rNtTPL1–6, arrowhead) specifically bound to probes 2 and 3 but not to 1.
Effects of overexpression of TOPLESS-like proteins on the responsiveness of BY-2 cells and tobacco plants to caryophyllene oxide
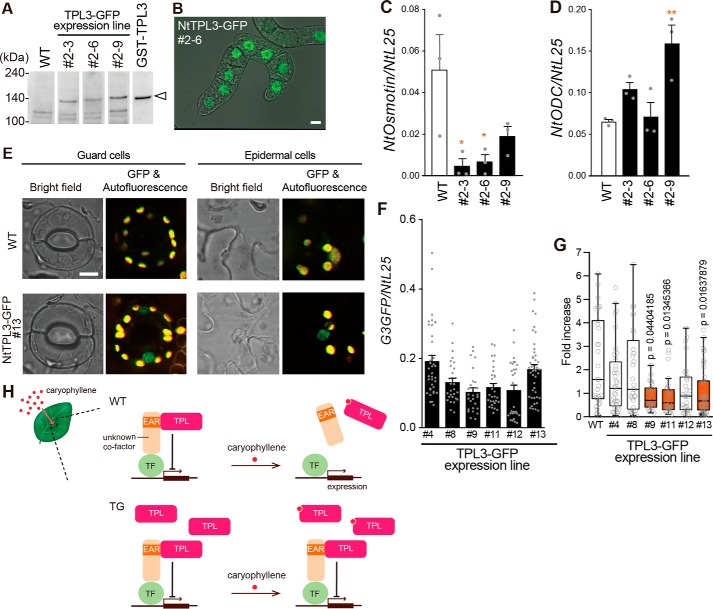
Recent studies suggest that TPL and TPL-related (TPR) proteins are transcriptional co-repressors (18, 19). Therefore, NtTPLs may be involved in the induction of NtOsmotin as transcriptional regulators as well as caryophyllene receptors. To examine whether NtTPLs are indeed receptors for caryophyllene and that the interaction affects NtOsmotin expression, we overexpressed NtTPLs in BY-2 cells and tobacco plants. First, we generated transgenic BY-2 cell lines expressing GFP-fusion NtTPL3 (Fig. 5A). The fusion proteins were localized to the nucleus in all cell lines (Fig. 5B). The nuclear localization of the NtTPL3 was consistent with its putative role as a transcriptional regulator. After 6 h of incubation with caryophyllene oxide, NtOsmotin expression was compared for WT and transgenic BY-2 cell lines. NtOsmotin expression in some transgenic lines was lower than in WT BY-2 cells (Fig. 5C). This result suggests that the interaction of NtTPL3 and caryophyllene oxide is involved in regulation of NtOsmotin expression. In contrast, NtODC expression after MeJA treatment was unchanged or significantly higher in the transgenic lines (Fig. 5D). Next, we generated transgenic tobacco plants expressing GFP fusion NtTPL3. The fusion proteins were also localized to the nucleus in tobacco (Fig. 5E). Also, we show the data demonstrating the expression level of NtTPL3-G3GFP in six transgenic lines (Fig. 5F). After 8 h of immersion in caryophyllene oxide solution, NtOsmotin expression in three of six transgenic lines was lower than in WT plants (Fig. 5G). Although other transgenic lines showed just a tendency for reduction in caryophyllene-induced NtOsmotin expression, likely due to endogenous NtTPL, the results indicated that the interaction of NtTPLs and caryophyllene structural analogues results in regulation of NtOsmotin expression. One model is that caryophyllene enters tobacco plant cells, binds NtTPLs coupled with transcriptional factors, and releases NtTPLs that otherwise suppress NtOsmotin gene transcription (Fig. 5H).
Figure 5.
Effects of overexpression of TPL on the response to caryophyllene oxide in BY-2 cell and tobacco plant. A, Western blotting using WT and transgenic BY-2 cells. White arrowhead, NtTPL3-GFP protein. B, localization of NtTPL3-GFP in transgenic BY-2 cells. Scale bar, 10 μm. C, dose-dependent induction of NtOsmotin in NtTPL3-GFP–overexpressing BY-2 cells. Caryophyllene oxide (1000 μm) or DMF (0.1%) as a control was applied to BY-2 cells for 6 h. The data for WT is the same as in Fig. 2B. Error bars, S.E. (n = 3). Statistics were calculated using one-way ANOVA and post hoc test (Bonferroni's test): *, p < 0.05. D, induction of NtODC in NtTPL3-GFP–overexpressing BY-2 cells. MJ (100 μm) or DMF (0.1%) was applied to BY-2 cells for 6 h. Error bars, S.E. (n = 3). Statistics were calculated using one-way ANOVA and post hoc test (Bonferroni's test): **, p < 0.01. E, localization of NtTPL3-GFP in transgenic tobacco leaves. Scale bar, 10 μm. F, expression level of GFP in NtTPL3-GFP–expressing plants. G, induction of NtOsmotin in NtTPL3-GFP–overexpressing plants. Leaves were immersed in caryophyllene oxide (300 μm) or DMF solution (0.1%) for 8 h. NtOsmotin expression is shown as -fold increase (n = 27–45). Whiskers, 1.5 times the interquartile range. Statistics were calculated using the Smirnov–Grubbs test, Kruskal–Wallis test, and post hoc test (Steel test). H, “one” model for control of NtOsmotin expression by caryophyllene.
Discussion
Previous studies concerning plant volatile reception have mainly focused on VOC-mediated output phenomena and ecological significance, but not on the molecular basis of VOC recognition. One study reported that mutations in transcriptional factors wrky40 and wrky6 affect (E)-2-hexenal–induced gene expression in Arabidopsis (20), suggesting the involvement of a transcriptional factor. In this study, we reported that a transcriptional co-repressor bound to VOCs, leading to gene regulation in plant cells. We provided several lines of evidence that support the hypothesis that NtTPLs are involved in sensing caryophyllene analogs (i.e. that these molecules have a specific structure–activity relationship (Figs. 1B and 3B), binding (Fig. 4), dose-dependent responses (Figs. 2B and 5C), and effects of overexpression in vitro and in vivo (Fig. 5)).
TPL/TPR proteins are known to be involved in regulation of gene repression as responses to hormone and stress in various developmental processes. For example, TPL/TPRs act as co-repressors for JA-mediated signaling by complexing with adaptor proteins such as JAZ5–JAZ8 or NINJA to which a transcription factor binds (21). It was recently reported that poplar TPR4 interacts with an effector protein from pathogenic fungus (22), suggesting that a member of the TPL/TPR proteins is functioning as a detector for an external cue in addition to the function as a transcriptional regulator. In this regard, our finding supports the dual function of TPL/TPRs such that they play a critical role not only in regulating gene expression as co-repressors, but also in binding individual VOCs. Given evolutionary conservation of TPL/TPR proteins (23, 24), it is also a crucial question whether their function in volatile detection is preserved. If so, it is also an intriguing question how TPL/TPR proteins modulate specific gene expression depending on each target molecule.
Even though it is possible that TPL's role is further upstream of the transcription factor, the fact that a transcription co-repressor functions as a VOC-binding protein is reminiscent of the recognition of plant hormones such as auxin and JA or of steroid hormones in mammals to regulate gene expression. The degradation of auxin/IAA and the JAZ complex causes activation of auxin- or JA-responsive genes (21). In animals, nuclear receptors, which comprise a large family of transcription factors (25), regulate gene expression, and among them, mall heterodimer partner contains a putative ligand-binding domain but lacks a DNA-binding domain (25, 26), regulating gene expression as a co-repressor (27). Although the caryophyllene–TPL pair is only the first case so far, and thus we cannot generalize the current finding for all VOC sensing, it is intriguing that plants appear to have evolved a VOC-sensing mechanism using nuclear proteins rather than membrane receptors such as odorant receptors in animals. This possibility is consistent with the observation that the number of transcription factor genes and the ratio of transcription factor genes to the total number of genes in plants are much higher than in animals (28, 29). In addition, the number of transcription factors has been expanded through evolutionary transitions in land plants (30).
Several questions remain to be addressed. First, the mechanism of caryophyllene and other VOC uptake by plant cells is unknown. It could be similar to the action of steroid hormones that enter a cell due to their hydrophobic characteristics, or it could be mediated by transporters in a way similar to VOC emission (31, 32). Second, the amounts of VOCs utilized are rather high. This concentration problem has been discussed in essentially all previous plant-volatile sensing studies. Even in mammals, the parts per thousand level of odorant perception cannot be explained by a micromolar level of odorant receptor sensitivity. There must be a mechanism such as uptake by transporters for establishing a high local concentration (33). Third, it takes several hours to observe VOC-induced gene expression in plant cells. Unlike an acute response in mammalian olfaction, the benefit of this slow response has to be investigated. Fourth, we did not demonstrate the necessity of TPL/TPR proteins to response caryophyllenes because of their functional redundancy.
Regardless of these important questions and the lack of a loss-of-function experiment, which was left to future studies due to technical limitation, the discovery of a VOC-binding protein in tobacco has shed light on the mechanisms of “olfaction” in plants. Now that a target for VOC binding is identified, elucidation of the exact mechanism and pathway leading to changes in a transcription regulatory complex that affects gene expression is an interesting topic to be explored in the future.
Experimental procedures
VOCs and plant regulators
Chemical compounds were purchased from TCI (Tokyo, Japan). The compound solutions for analysis using BY-2 cells were prepared as 1 m stocks in N,N-dimethylmethanamide (DMF; Wako, Osaka, Japan) and then diluted to the indicated concentrations (containing 0.1% DMF) before each experiment.
BY-2 cell material and culture conditions
WT tobacco N. tabacum L. cv. Bright Yellow 2 (BY-2) cells and transgenic cell lines were cultivated under dark conditions at 27 °C and 130 rpm in a BR-42FL MR incubator shaker (TAITEC, Saitama, Japan). Weekly, 0.3 ml of each culture was transferred to 30 ml of LS liquid medium.
Vector construction and transformation of BY-2 cell
Full-length NtTPL3 was cloned into vector pENTR/D-TOPO (Life Technologies, Inc.) and then cloned into binary vector pGWB551 with a GFP reporter (34) using Gateway LR Clonase (Life Technologies). The full-length osmotin promoter was also cloned into binary vector pGWB559 with an RFP reporter (34). Each construct was electroporated into Agrobacterium tumefaciens LBA4404 (Life Technologies) using a preset protocol of the Gene Pulser Xcell electroporation system (Bio-Rad).
On day 3, 4 ml of BY-2 cells was transferred to dishes, and 100 μl of A. tumefaciens overnight culture containing either pGWB551 (Gateway binary vector for C-terminal fusion with G3GFP, CaMV35S promoter) or pGWB559 (Gateway vector for TagRFP, Osmotin promoter) was added. After 48 h of co-cultivation at 27 °C, BY-2 cells were washed with 3% sucrose solution and transferred onto LS agar plates containing both cefotaxime (final concentration, 250 μg/ml) and hygromycin (final concentration, 50 μg/ml). After 4 weeks, calli were transferred onto fresh LS agar plates and further incubated. After 1 week, several calli expressing GFP or RFP were selected using an LED light (OptoCode Corp.) and transferred onto fresh LS agar plates. After another week, to establish transgenic cell lines in liquid culture, a callus was transferred into 10 ml of LS liquid medium. After 3 days, the callus was fragmented using a 10-ml pipette. After another week following the 3 days, liquid cell cultures were examined under a confocal microscope to determine whether the fluorescent reporter protein was expressed. When cells had nearly grown to saturation in liquid culture, cultures were transferred into 95 ml of LS liquid medium. The growth rate of transgenic cell lines became the same as the WT cell line, so 0.3 ml of each culture was transferred to 30 ml of LS liquid medium weekly for passage.
Confocal imaging
Confocal images were captured using an IX-71 inverted microscope (Olympus, Tokyo, Japan) equipped with a CSU10 confocal scanner unit (Yokogawa Electric Corp., Tokyo, Japan). A water immersion ×60 objective lens (Olympus) was used for observation of GFP fusion protein, and a ×40 objective lens (Olympus) was used for RFP reporter assays. Images were processed with ImageJ software (National Institutes of Health).
Plant material and growth conditions
N. tabacum L. cv. Samsun-NN WT plants were grown under a 16/8-h light/dark cycle at 25 °C, 65% humidity in an LPH-220SP growth chamber (NK Systems, Osaka, Japan). Surface-sterilized seeds were sown on medium containing Murashige and Skoog salts, 1 mg/liter thiamine hydrochloride salts (Wako, Osaka, Japan), and 0.1 g/liter myo-inositol (Wako, Osaka, Japan) solidified with 0.8% agar BA-10 (INA, Nagano, Japan). Seedlings were grown in an Agripot container (Kirin Agribio, Shizuoka, Japan) and used for experiments 4 weeks after sowing.
Transformation of tobacco plants
N. tabacum L. cv. Petit Havana SR1 WT plants were grown under a 14/10-h light/dark cycle at 25 °C, 65% humidity in a growth chamber. Surface-sterilized seeds were sown on Murashige and Skoog medium plates. Seedlings were transplanted into a plant box 2 weeks after sowing. The upper three leaves (at 1 month) were cut in an A. tumefaciens 40-h culture (diluted 20 times with sterilized water) containing pGWB551 with NtTPL3.
After 3 days co-cultivation at 28 °C on Murashige and Skoog medium containing 0.1 mg/liter 1-naphthaleneacetic acid and 1 mg/liter 6-benzylaminopurine, leaves were transplanted on Murashige and Skoog medium containing 0.1 mg/liter 1-naphthaleneacetic acid and 1 mg/liter 6-benzylaminopurine, cefotaxime (final concentration, 200 μg/ml), and hygromycin (final concentration, 25 μg/ml) at 25 °C. After 2 months, plants were transplanted to Murashige and Skoog medium containing both cefotaxime (final concentration, 100 μg/ml) and hygromycin (final concentration, 25 μg/ml). After 1 month, plants were examined for expression of GFP by PCR. The primers used were 5′-GACACGTGCTGAAGTCAAGT-3′ and 5′-CCATGCCATGTGTAATCCCA-3′.
Exposure of tobacco to volatiles in Agripot containers
A 40-μl aliquot of each volatile compound was placed in an enclosed Agripot in which 4-week-old tobacco plants were grown on Murashige and Skoog agar medium for 8 h in the dark in an LPH-220SP growth chamber. Four independent plants harvested in a single experiment were pooled to prepare one RNA sample.
Treatment of tobacco in solution with volatiles
Tobacco leaves were harvested and immersed into 2 ml of Murashige and Skoog liquid medium containing caryophyllene structural analogues (final concentration, 1 mm) or DMF (final concentration, 0.1%) in 12-well plates (Sumitomo Bakelite, Tokyo, Japan). They were sealed and incubated under dark conditions in an LPH-220SP growth chamber.
Treatment of BY-2 cells with volatiles
BY-2 cells (day 3) were dispensed to 12-well plates (2 ml/well), and volatile compounds were added at the indicated concentrations for each experiment, which was performed in duplicate. They were sealed and incubated under dark conditions at 27 °C and 130 rpm on an NR-30 shaker (TAITEC) in an LPH-220SP growth chamber. After incubation, each culture was collected for RNA extraction.
RNA extraction
After incubation, samples were collected, frozen in liquid nitrogen, and homogenized with a Micro Smash MS-100R cell disruptor (TOMY, Tokyo, Japan) for RNA extraction. Total RNA was extracted from crushed leaves using a QIAshredder spin column and RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. After RQ1 RNase-free DNase treatment (Promega), total RNA was used for cDNA synthesis.
Q-PCR
Complementary DNA was synthesized using Superscript III reverse transcriptase (Life Technologies) and analyzed by Q-PCR. Q-PCR was performed using a StepOnePlus System (Thermo Fisher Scientific) with Fast SYBR Green Master Mix (Thermo Fisher Scientific).
The primers used were 5′-TTGAGGACAACAACACCCTTG-3′ and 5′-ACATCTTCTTCACGGCATC-3′ for ribosomal protein L25 (NtL25), 5′-TCCTCCTTGCCTTGGTGACTTATA-3′ and 5′-ATCACCCAAGTTTGGCCTCGATC-3′ for NtOsmotin, 5′-TAGAAGGCGGGACAACACAAC-3′ and 5′-TTTAACCATGTATCTGGGAATGGAC-3′ for acidic chitinase III (NtACIII), and 5′-TTCCGACGACTGTGTTTGG-3′ and 5′-AACCTGCAGCTCCGGTAAC-3′ for ornithine decarboxylase (NtODC). Expression of each gene was quantified by Q-PCR and normalized to NtL25.
Preparation of samples for GC/MS
BY-2 cells were incubated with (E)-β-caryophyllene, α-caryophyllene, or caryophyllene oxide (1 mm, final concentration) for 3 h. After incubation, cells were collected and washed three times. They were frozen and disrupted with a mortar and pestle. Disrupted cells were mixed with 2 ml of water (one-tenth volume of liquid medium) and 2 ml of ethyl acetate. To separate the ethyl acetate extract from the emulsion, the mixture was centrifuged. The ethyl acetate layer was collected to new vials. A 1-μl aliquot of each sample was injected to a GC/MS with an MS-E10 microsyringe (Ito Corp., Shizuoka, Japan). For preparation of airborne compounds in the headspace, caryophyllene structural analogs were placed in enclosed plant pots (40 μl/pot) for 8 h, and 10 μl of headspace was directly sampled using a gas-tight model 1701 N SYR syringe (Hamilton).
Chromatographic conditions for GC/MS
GC/MS on a GCMS-TQ8030 instrument with an OPTIC-4 inlet (Shimadzu, Kyoto, Japan) was used to examine the ethyl acetate extract. The compounds were separated using a Stabilwax column (60.0 m × 0.32-mm inner diameter, 0.5-μm film thickness). The column temperature was held at 40 °C for 2 min and then programmed to rise to 230 °C at 5 °C/min, and finally held at this temperature for 5 min. The interface temperature was maintained at 230 °C, and the ion source temperature was set at 230 °C. Peak identities were confirmed by matching the component mass spectra to the National Institute of Standards and Technology Mass Spectral Database and by matching the retention time and mass spectra of the peaks to the data observed for the pure compounds. Mass spectra (20.0–300.0 m/z) were obtained in full scan mode. Products were analyzed quantitatively by comparison with the peak area of compounds in total ion chromatograms.
Preparation of protein extracts from leaves
Leaves were frozen in liquid nitrogen and homogenized with an MS-100R Micro Smash disruptor. Total protein was extracted from crushed leaves in an extraction buffer containing 50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 5 mm KCl, 1 mm DTT, 10 μm MG132 proteasome inhibitor (Merck), and the cOmplete, Mini, EDTA-free protease inhibitor mixture tablet (Roche Applied Science) according to the supplier's instructions. After centrifugation (10,000 × g for 10 min at 4 °C), the supernatant was collected and immediately used for a pulldown assay. Total soluble protein concentration was measured by a Bradford assay on a microplate absorbance reader (Bio-Rad).
Preparation of recombinant NtTPL proteins
The cDNA sequences encoding NtTPLs were identified using 5′- and 3′-RACE with a SMARTer RACE cDNA amplification kit (Clontech). The NtTPL sequences were inserted into vector pGEX-6P-1 (GE Healthcare). The expression construct was transformed into the expression host Escherichia coli Rosetta 2(DE3) (Novagen). After preculture at 37 °C to the mid-log phase, expression of the target GST fusion proteins was induced by adding isopropyl 1-thio-β-d-galactopyranoside to 0.1 mm final concentration at 25 °C for 4 h before harvest. Pellets were resuspended in a buffer containing 50 mm Tris-HCl (pH 7.4), 500 mm NaCl, 5 mm KCl, 10 mm DTT, 1% Triton X-100, and a cOmplete, Mini, EDTA-free protease inhibitor mixture tablet (Roche Applied Science) according to the supplier's instructions and then sonicated on ice. After centrifugation (10,000 × g for 5 min at 4 °C), the supernatants were collected. They were purified using GSH Sepharose 4B medium (GE Healthcare) and immediately used for a pulldown assay. The concentration of purified recombinant proteins was measured by Bradford assay on a Bio-Rad microplate absorbance reader.
Pulldown assay
Total protein extracts (1 mg) of N. tabacum leaves or recombinant NtTPL proteins (10 μg) were incubated with (E)-β-caryophyllene derivatives or propylamine-linked beads (10 μl) for 1 h at 4 °C. After incubation, beads were separated by a magnet (Veritas) from the supernatant and washed three times with protein extraction buffer containing 50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 5 mm KCl, 1 mm DTT, 10 μm MG132 proteasome inhibitor (Merck), and the cOmplete, Mini, EDTA-free protease inhibitor mixture tablet (Roche Applied Science) according to the supplier's instructions. 0.1% of input sample and 25% of extracts from each bead were loaded per lane. SDS-PAGE and silver staining were carried out to confirm the size of the proteins and their binding activities. Samples stained without glutaraldehyde were in-gel digested with trypsin and analyzed by LC/tandem MS (LC-MS/MS).
Chromatographic conditions for LC-MS/MS
LC-MS/MS (EASY-nLC with Q Exactive system, Thermo Fisher Scientific) was used to examine binding proteins after in-gel digestion. The peptides were separated using C18 columns for a trap column (SC001, 20 mm × 100-μm inner diameter, Thermo Fisher Scientific) and analytical column (NTCC-360/75-3-125, 15 cm × 100-μm inner diameter, Nikkyo Technos). Water (solvent A) and acetonitrile (solvent B) containing 0.1% (v/v) formic acid were used as mobile phase. The gradient was programmed to rise from 0 to 35% B during the first 20 min, from 35 to 100% B for 2 min, and finally held at 100% B for 8 min at a flow rate of 300 nl/min. Fragments were detected by electrospray ionization in positive ion mode. Mass spectra (200–2000 m/z) were obtained in full MS and ddMS2 scan modes. Data were acquired using Xcalibur software (Thermo Fisher Scientific). Data were analyzed using Proteome Discoverer (Thermo Fisher Scientific), PEAKS (Bioinformatics Solutions Inc.), and Mascot software (Matrix Science).
Preparation of antibody
Anti-NtTPL3 antibody was generated (Evebio Science, Wakayama, Japan) against a keyhole limpet hemocyanin–conjugated synthetic peptide (C+DNGILNGRTASSS, amino acids 1045–1057 from the C-terminal region of NtTPL3; Scrum Inc., Tokyo, Japan). Specific antibody was purified from immunized rabbit serum using a SulfoLink immobilization kit for peptides (Thermo Fisher Scientific).
Western blotting
The proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (PALL). After blocking with 5% skim milk, the membrane was incubated with anti-NtTPL3 antibody (1:100). The primary antibody was immunoblotted with horseradish peroxidase–conjugated anti-rabbit secondary antibody (1:5000) and detected with ImmunoStar LD reagent (Wako, Osaka, Japan).
Synthesis of (E)-β-caryophyllene oxime derivative with amino group for biotinylation (see Fig. S3A)
A solution of caryophyllene oxide (15 g, 68 mmol) in ethyl acetate (400 ml) was ventilated with ozone from an ON-3-2 ozone generator (Nippon Ozone Co., Ltd.) at 0 °C for 2 h. To the reaction mixture was added dimethyl sulfide (80 ml) at room temperature overnight, and then it was poured into water and extracted with ethyl acetate. After concentration, silica gel chromatography gave ketone (1) as a colorless oil (9.8 g, 65%). A solution of 1 (9.0 g, 40 mmol) in 99% EtOH (600 ml) was added to a Zn–Cu couple (140 g) and refluxed overnight. After filtration through Celite and concentration, the sample was resuspended in diethyl ether. After filtration through Celite and concentration again, silica gel chromatography gave ketone (2) as a colorless oil (6.8 g, 81%). To a solution of 2 (1.5 g, 7.2 mmol) in benzene (20 ml) was added 3-aminooxypropyl-1-azide (1.0 g, 8.6 mmol) and acetic acid (1 ml), and the mixture was stirred at room temperature for 4 h (35). After concentration, silica gel chromatography gave oxime (3) as a colorless oil (2.0 g, 90%). To a solution of 3 (135 mg, 0.44 mmol) in tetrahydrofuran (THF; 5 ml) was added H2O (100 μl) and triethylphosphine in toluene (20%, 355 μl, 0.59 mmol), and the mixture was stirred at room temperature for 1.5 h. After concentration, silica gel chromatography gave amine (4) as a colorless oil (122 mg, 99%).
Synthesis of (E)-β-caryophyllene derivative with three-membered ring with amino group for biotinylation (see Fig. S3B)
To [(CH3COO)2Rh]2·2H2O (50 mg) in CH2Cl2 (20 ml) was added ethyl diazoacetate (0.5 ml, 4.7 mmol), CH2Cl2 (10 ml), and (E)-β-caryophyllene (1.5 ml, 6.6 mmol), and the mixture was stirred under argon at room temperature overnight (36). After concentration, silica gel chromatography gave ether (5) as a colorless oil (381 mg, 28% and 345 mg, 26%; separate syntheses). To a solution of 5 (381 mg, 1.3 mmol and 345 mg, 1.2 mmol; separate syntheses) in THF (9 ml), H2O (6 ml), and MeOH (6 ml) was added LiOH·H2O (230 mg) at 80 °C for 4.5 h. The reaction mixture was adjusted with 3 n HCl to pH 3 and extracted with ethyl acetate. After concentration, silica gel chromatography gave carboxylic acid (6) as a brown oil (127 mg, 37% and 186 mg, 59%; separate syntheses). To a solution of 6 (205 mg, 0.78 mmol) in benzene (4 ml) was added SOCl2 (300 μl, 4.1 mmol) under argon at 80 °C for 4 h. The reaction product, carboxylic acid chloride (7) in dioxane (14 ml), was treated with NH3 at room temperature for 15 min. After filtration through Celite, silica gel chromatography gave amide (8) (122 mg, 49%). A solution of 8 (122 mg, 0.38 mmol) in THF (12 ml) was added to lithium aluminum hydride (80 mg, 0.26 mmol) under argon at 75 °C for 3 h and then treated with H2O and NaOH. After filtration through Celite and concentration, silica gel chromatography gave amine (9) as a colorless oil (75 mg, 79%).
Preparation of probes for a pulldown assay
(E)-β-Caryophyllene derivatives with amino group and propylamine (as control) were biotinylated by Biotin-XX, SE (6-((6-((biotinoyl)amino)hexanoyl)amino)hexanoic acid, succinimidyl ester) (Thermo Fisher Scientific), and 1 nmol of biotinylated ligand was immobilized per mg (= 100 μl) of streptavidin-coated beads (Dynabeads M-280 Streptavidin, VERITAS).
Phylogenetic analysis
S. lycopersicum TPL was used as query sequence for BLAST searches on the Sol Genomics Network (https://solgenomics.net).6 Candidates that were predicted by PROSITE (http://prosite.expasy.org)6 to have a LisH domain, CLISH domain, and WD40 repeats were defined as TPL proteins. Amino acid sequences of TPL proteins were aligned by MAFFT version 7 software (http://mafft.cbrc.jp/alignment/server/).6 MEGA7 software generated phylogenetic trees with the neighbor-joining method and Jones-Taylor-Thornton matrix-based method. The percentages of replicate trees in which associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches.
Statistics
Statistical analyses were performed with EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan) (37), which is a graphical user interface for R version 3.3.3 (38, 39). More precisely, it is a modified version of R commander version 2.3-2 designed to add functions frequently used in biostatistics.
Author contributions
A. N., S. Hosokawa, and K. T. designed the study. A. N. provided data, except imaging, identified NtTPLs as binding proteins for volatile caryophyllene analogs, synthesized biotinylated caryophyllene derivatives with help from K. I. and H. W., and generated transgenic BY-2 cell lines with help from T. H. and S. Hasezawa. K. M. and T. H. performed imaging experiments. A. N. generated vector, and T. K. and K. M. generated transgenic tobacco using it. A. N. and K. T. wrote the paper with substantial contributions from other authors.
Supplementary Material
Acknowledgments
We thank M. Suzuki for providing seeds of N. tabacum L. cv. Samsun-NN, and we thank K. Sugimoto, H. Satsu, Y. Tada, H. Iida, J. Nabekura, T. Higashiyama, T. Tashiro, S. Nagata, S. Arimura, and members of the Touhara laboratory for valuable discussions.
This work was supported in part by the ERATO Touhara Chemosensory Signal Project from the Japan Science and Technology Agency (JST), Japan (Grant JPMJER1202) and by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to K. T.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- VOC
- volatile organic compound
- GPCR
- G protein–coupled receptor
- MeJA
- methyl jasmonate
- SA
- salicylic acid
- Q-PCR
- quantitative PCR
- RFP
- red fluorescent protein
- TPL
- TOPLESS-like protein
- TPR
- TPL-related
- DMF
- N,N-dimethylmethanamide
- THF
- tetrahydrofuran
- RACE
- rapid amplification of cDNA ends.
References
- 1. Farmer E. E., and Ryan C. A. (1990) Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. U.S.A. 87, 7713–7716 10.1073/pnas.87.19.7713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heil M., and Karban R. (2010) Explaining evolution of plant communication by airborne signals. Trends Ecol. Evol. 25, 137–144 10.1016/j.tree.2009.09.010 [DOI] [PubMed] [Google Scholar]
- 3. Kessler A., and Baldwin I. T. (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291, 2141–2144 10.1126/science.291.5511.2141 [DOI] [PubMed] [Google Scholar]
- 4. Shulaev V., Silverman P., and Raskin I. (1997) Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385, 718–721 10.1038/385718a0 [DOI] [Google Scholar]
- 5. Arimura G., Ozawa R., Shimoda T., Nishioka T., Boland W., and Takabayashi J. (2000) Herbivory-induced volatiles elicit defence genes in lima bean leaves. Nature 406, 512–515 10.1038/35020072 [DOI] [PubMed] [Google Scholar]
- 6. Ton J., D'Alessandro M., Jourdie V., Jakab G., Karlen D., Held M., Mauch-Mani B., and Turlings T. C. (2007) Priming by airborne signals boosts direct and indirect resistance in maize. Plant J. 49, 16–26 [DOI] [PubMed] [Google Scholar]
- 7. Runyon J. B., Mescher M. C., and De Moraes C. M. (2006) Volatile chemical cues guide host location and host selection by parasitic plants. Science 313, 1964–1967 10.1126/science.1131371 [DOI] [PubMed] [Google Scholar]
- 8. Taddese B., Upton G. J., Bailey G. R., Jordan S. R., Abdulla N. Y., Reeves P. J., and Reynolds C. A. (2014) Do plants contain G protein-coupled receptors? Plant Physiol. 164, 287–307 10.1104/pp.113.228874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eichinger L., Pachebat J. A., Glöckner G., Rajandream M. A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Tunggal B., Kummerfeld S., Madera M., Konfortov B. A., Rivero F., et al. (2005) The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 10.1038/nature03481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagata T., Nemoto Y., and Hasezawa S. (1992) Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int. Rev. Cytol. 132, 1–30 10.1016/S0074-7696(08)62452-3 [DOI] [Google Scholar]
- 11. De Moraes C. M., Mescher M. C., and Tumlinson J. H. (2001) Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature 410, 577–580 10.1038/35069058 [DOI] [PubMed] [Google Scholar]
- 12. Delphia C. M., Mescher M. C., and De Moraes C. M. (2007) Induction of plant volatiles by herbivores with different feeding habits and the effects of induced defenses on host-plant selection by thrips. J. Chem. Ecol. 33, 997–1012 10.1007/s10886-007-9273-6 [DOI] [PubMed] [Google Scholar]
- 13. Subramanyam K., Arun M., Mariashibu T. S., Theboral J., Rajesh M., Singh N. K., Manickavasagam M., and Ganapathi A. (2012) Overexpression of tobacco osmotin (Tbosm) in soybean conferred resistance to salinity stress and fungal infections. Planta 236, 1909–1925 10.1007/s00425-012-1733-8 [DOI] [PubMed] [Google Scholar]
- 14. Imanishi S., Hashizume K., Nakakita M., Kojima H., Matsubayashi Y., Hashimoto T., Sakagami Y., Yamada Y., and Nakamura K. (1998) Differential induction by methyl jasmonate of genes encoding ornithine decarboxylase and other enzymes involved in nicotine biosynthesis in tobacco cell cultures. Plant Mol. Biol. 38, 1101–1111 10.1023/A:1006058700949 [DOI] [PubMed] [Google Scholar]
- 15. Lawton K., Ward E., Payne G., Moyer M., and Ryals J. (1992) Acidic and basic class III chitinase mRNA accumulation in response to TMV infection of tobacco. Plant Mol. Biol. 19, 735–743 10.1007/BF00027070 [DOI] [PubMed] [Google Scholar]
- 16. Rasmann S., Köllner T. G., Degenhardt J., Hiltpold I., Toepfer S., Kuhlmann U., Gershenzon J., and Turlings T. C. (2005) Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434, 732–737 10.1038/nature03451 [DOI] [PubMed] [Google Scholar]
- 17. Degenhardt J., Hiltpold I., Köllner T. G., Frey M., Gierl A., Gershenzon J., Hibbard B. E., Ellersieck M. R., and Turlings T. C. (2009) Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proc. Natl. Acad. Sci. U.S.A. 106, 13213–13218 10.1073/pnas.0906365106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pauwels L., Barbero G. F., Geerinck J., Tilleman S., Grunewald W., Pérez A. C., Chico J. M., Bossche R. V., Sewell J., Gil E., García-Casado G., Witters E., Inzé D., Long J. A., De Jaeger G., et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 10.1038/nature08854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Szemenyei H., Hannon M., and Long J. A. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 10.1126/science.1151461 [DOI] [PubMed] [Google Scholar]
- 20. Mirabella R., Rauwerda H., Allmann S., Scala A., Spyropoulou E. A., de Vries M., Boersma M. R., Breit T. M., Haring M. A., and Schuurink R. C. (2015) WRKY40 and WRKY6 act downstream of the green leaf volatile E-2-hexenal in Arabidopsis. Plant J 83, 1082–1096 10.1111/tpj.12953 [DOI] [PubMed] [Google Scholar]
- 21. Pérez A. C., and Goossens A. (2013) Jasmonate signalling: a copycat of auxin signalling? Plant Cell Environ. 36, 2071–2084 10.1111/pce.12121 [DOI] [PubMed] [Google Scholar]
- 22. Petre B., Saunders D. G., Sklenar J., Lorrain C., Win J., Duplessis S., and Kamoun S. (2015) Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Mol. Plant Microbe Interact. 28, 689–700 10.1094/MPMI-01-15-0003-R [DOI] [PubMed] [Google Scholar]
- 23. Causier B., Ashworth M., Guo W., and Davies B. (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158, 423–438 10.1104/pp.111.186999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Causier B., Lloyd J., Stevens L., and Davies B. (2012) TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signal Behav. 7, 325–328 10.4161/psb.19283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gronemeyer H., Gustafsson J. A., and Laudet V. (2004) Principles for modulation of the nuclear receptor superfamily. Nat. Rev. Drug Discov. 3, 950–964 10.1038/nrd1551 [DOI] [PubMed] [Google Scholar]
- 26. Seol W., Choi H. S., and Moore D. D. (1996) An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science 272, 1336–1339 10.1126/science.272.5266.1336 [DOI] [PubMed] [Google Scholar]
- 27. Zou A., Lehn S., Magee N., and Zhang Y. (2015) New insights into orphan nuclear receptor SHP in liver cancer. Nucl. Receptor Res. 2, 101162 10.11131/2015/101162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O. J., Samaha R. R., Creelman R., Pilgrim M., Broun P., Zhang J. Z., Ghandehari D., et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110 10.1126/science.290.5499.2105 [DOI] [PubMed] [Google Scholar]
- 29. Libault M., Joshi T., Benedito V. A., Xu D., Udvardi M. K., and Stacey G. (2009) Legume transcription factor genes: what makes legumes so special? Plant Physiol. 151, 991–1001 10.1104/pp.109.144105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Banks J. A., Nishiyama T., Hasebe M., Bowman J. L., Gribskov M., dePamphilis C., Albert V. A., Aono N., Aoyama T., Ambrose B. A., Ashton N. W., Axtell M. J., Barker E., Barker M. S., Bennetzen J. L., et al. (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 10.1126/science.1203810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Widhalm J. R., Jaini R., Morgan J. A., and Dudareva N. (2015) Rethinking how volatiles are released from plant cells. Trends Plant Sci. 20, 545–550 10.1016/j.tplants.2015.06.009 [DOI] [PubMed] [Google Scholar]
- 32. Adebesin F., Widhalm J. R., Boachon B., Lefèvre F., Pierman B., Lynch J. H., Alam I., Junqueira B., Benke R., Ray S., Porter J. A., Yanagisawa M., Wetzstein H. Y., Morgan J. A., Boutry M., et al. (2017) Emission of volatile organic compounds from petunia flowers is facilitated by an ABC transporter. Science 356, 1386–1388 10.1126/science.aan0826 [DOI] [PubMed] [Google Scholar]
- 33. Matsui K. (2016) A portion of plant airborne communication is endorsed by uptake and metabolism of volatile organic compounds. Curr. Opin. Plant Biol. 32, 24–30 10.1016/j.pbi.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 34. Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., Maeo K., Tabata R., Kawai T., Tanaka K., Niwa Y., Watanabe Y., Nakamura K., Kimura T., and Ishiguro S. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100 10.1271/bbb.70216 [DOI] [PubMed] [Google Scholar]
- 35. Ki S. W., Ishigami K., Kitahara T., Kasahara K., Yoshida M., and Horinouchi S. (2000) Radicicol binds and inhibits mammalian ATP citrate lyase. J. Biol. Chem. 275, 39231–39236 10.1074/jbc.M006192200 [DOI] [PubMed] [Google Scholar]
- 36. Muller P., and Tohill S. (2000) Intermolecular cyclopropanation versus CH insertion in Rh-II-catalyzed carbenoid reactions. Tetrahedron 56, 1725–1731 10.1016/S0040-4020(00)00076-4 [DOI] [Google Scholar]
- 37. Kanda Y. (2013) Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 48, 452–458 10.1038/bmt.2012.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. RCore Team (2017) R, a Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 39. Komsta L. (2011) Tests for outliers. R package version 0.14, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.