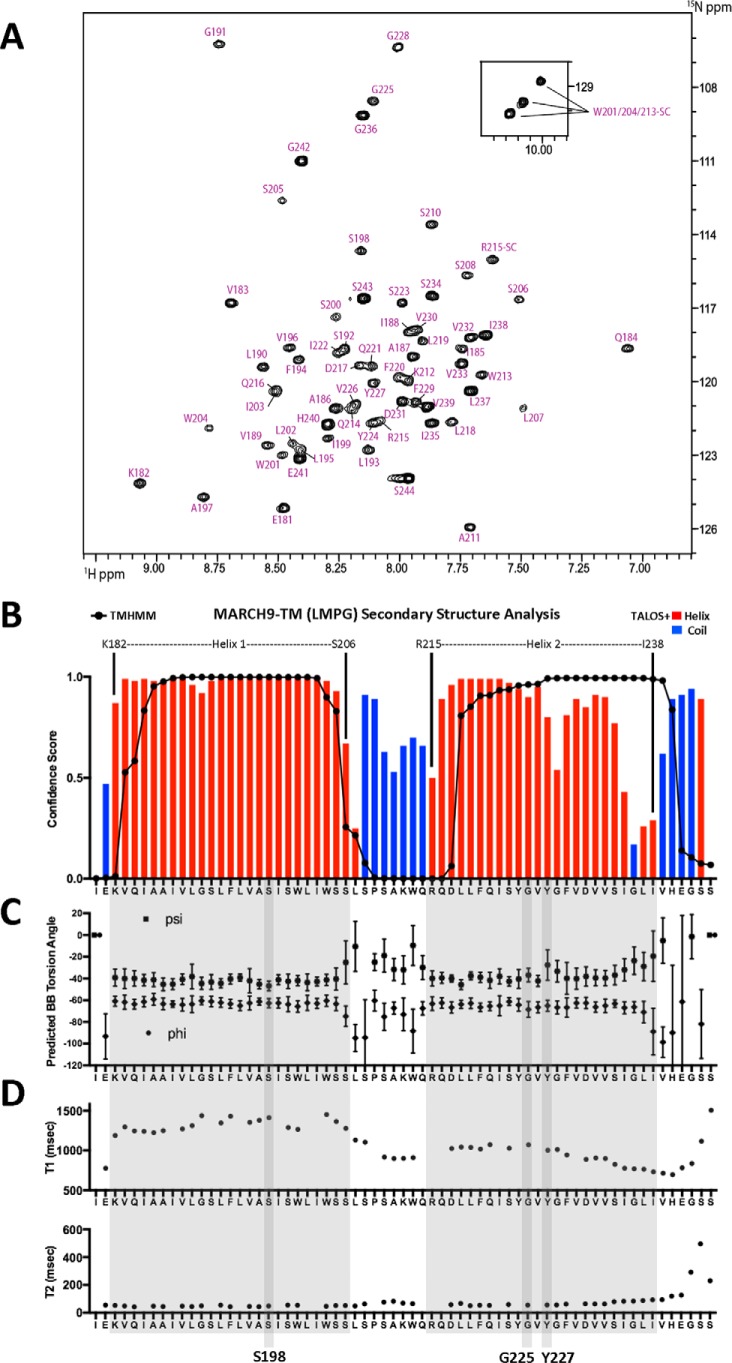
Figure 8.
Structural analysis of a MARCH9 TM–loop–TM fragment by solution NMR. A, assigned 1H–15N HSQC spectrum of a uniformly 15N,13C,2H 80%-labeled, 65-amino acid MARCH9 TM–loop–TM peptide (1 mm) in 250 mm LMPG, 20 mm phosphate buffer, pH 6.8, 5% D2O at 600 MHz and 40 °C. The TM–loop–TM peptide contained mutations C223S, M226V, M230V, and C234S to aid in production and an I239V substitution that is present in the mouse sequence (see “Experimental procedures”). B, secondary structure prediction from assigned backbone chemical shifts using TALOS+ (49). Confidence scores for residues predicted to be part of a helix (red bars) or random coil (blue bars) are superimposed with TMHMM predictions (black lines/dots) for comparison. C, backbone torsion angle ϕ (dots) and ψ (triangles) predictions from TALOS+. Values are in degrees, and error bars represent the estimated S.D. of the prediction error. D, backbone 15N T1 (top) and T2 (bottom) relaxation times (milliseconds) determined for a uniformly 15N-labeled MARCH9–TM sample prepared as in A and analyzed at 600 MHz and 40 °C. Helix 1 and Helix 2 are highlighted in large shaded boxes. The positions of key residues Ser-198, Gly-225, and Tyr-227 are highlighted in vertical shaded boxes.