Abstract
The p63 transcription factor, a p53 family protein, regulates genes involved in various cellular processes, including cell growth and differentiation. We previously showed that RNA-binding motif protein (Rbm38) is a p63 target and, in turn, regulates p63α mRNA stability by binding to the AU/U-rich element in its 3′UTR. Interestingly, Rbm38 can be phosphorylated at serine 195, altering its ability to regulate mRNA translation. However, whether the Ser-195 phosphorylation affects Rbm38's ability to destabilize p63 mRNA remains unclear. Here, using MCF7 and HaCaT cells, we showed that ectopic expression of phosphomimetic Rbm38-S195D increases, whereas WT Rbm38 and nonphosphorylatable Rbm38-S195A decrease p63α protein and transcript levels. We also found that upon activation of glycogen synthase kinase 3β (GSK3β), phosphorylation of Rbm38 at Ser-195 is increased, enhancing p63α expression in an Rbm38-dependent manner. To confirm this, we generated mouse embryo fibroblasts (MEFs) in which Ser-193 in mouse Rbm38 (equivalent to Ser-195 in human Rbm38) was substituted with aspartic acid (Rbm38S193D/S193D) or alanine (Rbm38S193A/S193A). We observed that the p63 transcript level was increased in Rbm38S193D/S193D MEFs, but decreased in Rbm38S193A/S193A MEFs. Mechanistically, we found that WT Rbm38, but not Rbm38-S195D, is required for p63 mRNA degradation mediated by microRNA 203 (miR203). Furthermore, we noted that Argonaute 2 (Ago2), a key regulator in microRNA-mediated mRNA decay, associates with WT Rbm38, and this association was reduced by Ser-195 phosphorylation. Together, our results reveal a critical mechanism by which Ser-195 phosphorylation in Rbm38 increases p63 expression by attenuating the association of Rbm38 with the Ago2–miR203 complex.
Keywords: p63, protein phosphorylation, transcription factor, post-transcriptional regulation, microRNA (miRNA), Argonaute 2, mRNA stability, RNA-binding motif protein 38 (Rbm38)
Introduction
p63 belongs to the p53 family of transcription factors that include p53, p63, and p73 (1–3). Because of the usage of two distinct promoters, p63 is expressed as two major isoforms, called TAp63 and ΔNp63, both of which have multiple variants through alternative splicing at the C terminus (2). Like p53, p63 functions as a transcription factor and induces many targets involved in various cellular processes, including cell proliferation, differentiation, epithelial development, and cellular senescence (4–6). Interestingly, although p63 shares many functional properties with p53, p63 is not a classic tumor suppressor as p63 is rarely mutated in human cancers. Indeed, p63 exhibits unique functions in the regulation of differentiation and development (6). Consistently, p63-deficient mice exhibit severe developmental defects in limb, skin, hair, teeth, and mammary and salivary glands (5, 6). Thus, exploring how p63 is regulated is critical for understanding the biological function of p63.
The RNA-binding protein Rbm38, also called RNPC1, is a target of the p53 family, including p63, as well as E2F1 (7, 8). Rbm38 is found to regulate expression of genes necessary for several cellular processes, including cell growth and differentiation. For example, Rbm38 is found to regulate Mdm2, p21, p73, and Pten mRNA stability through binding to the AU/U-rich elements in their 3′UTRs (8–11). We also found that Rbm38 decreases p63α/β mRNA stability by binding to AU/U-rich elements in their 3′UTR (12, 13). By contrast, Rbm38 enhances p63γ mRNA stability by binding to a GU-rich element in p63γ 3′UTR, which has a sequence different from that in p63α/β 3′UTR (14). In addition, we found that Rbm38 interacts with p53 mRNA and represses p53 translation (15). Moreover, Rbm38 is capable of regulating gene expression by modulating miRNA2 activity (16).
GSK3β is a serine/threonine kinase that is required for multiple cellular functions, including metabolism, cell growth, and differentiation (17). Previous studies showed that GSK3β can phosphorylate β-catenin and promotes its degradation (18, 19). Our group also showed that Rbm38 can be phosphorylated by GSK3β at Ser-195, and Ser-195 phosphorylation converts Rbm38 from a repressor to an activator of p53 mRNA translation via altered interaction with eIF4E (20). The unique function of Ser-195 phosphorylation in Rbm38 let us postulate whether Ser-195 phosphorylation alters the ability of Rbm38 to regulate p63 mRNA stability. Indeed, we found that Ser-195 phosphorylation abrogates the ability of Rbm38 to decrease p63α expression. Furthermore, we showed that WT Rbm38, but not phosphomimetic Rbm38-S195D, physically associates with Ago2, which is required for miR203 to target p63α mRNA for degradation. Together, we uncovered a novel mechanism by which Ser-195 phosphorylation in Rbm38 increases p63 expression by attenuating the association of Rbm38 with the Ago2–miR203 complex.
Results
Ser-195 phosphorylation alters the ability of Rbm38 to inhibit p63 mRNA stability
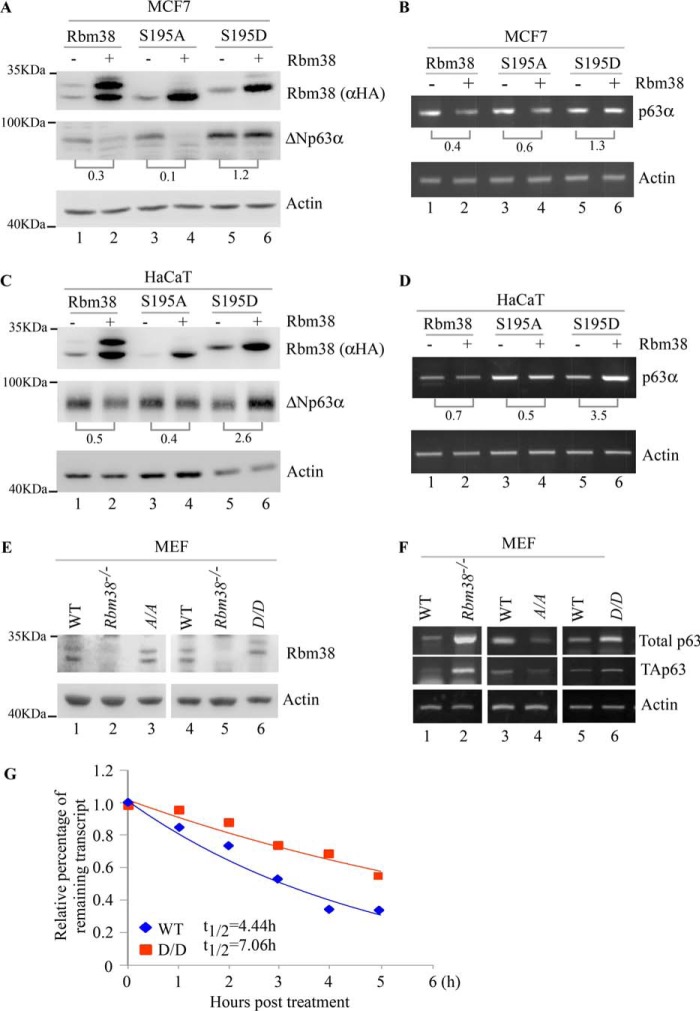
Previously, we found that Rbm38 negatively regulates p63α mRNA stability through binding to the AU/U-rich elements in p63 3′UTR (12, 13). We also found that phosphorylation of Ser-195 in Rbm38 alters the ability of Rbm38 from a repressor to an activator of p53 mRNA translation (20). Thus, we wanted to determine whether Ser-195 phosphorylation alters the ability of Rbm38 to regulate p63 mRNA stability. To test this, p63 expression was examined in multiple cell lines that inducibly express HA-tagged WT Rbm38, nonphosphorylatable Rbm38-S195A, or phosphomimetic Rbm38-S195D. We would like to mention that Rbm38 protein is expressed as two bands in the SDS-polyacrylamide gel: the upper (slow-migrating) band representing Ser-195–phosphorylated Rbm38; the lower (fast-migrating) band representing unphosphorylated Rbm38 (8, 10, 13, 15, 20). Consistently, we found that upon induction, Rbm38 protein was expressed as two bands in cell lines that inducibly express HA-tagged WT Rbm38 (Fig. 1, A and C, lanes 1 and 2), whereas Rbm38-S195A protein was expressed as the fast-migrating nonphosphorylatable band (Fig. 1, A and C, lanes 3 and 4). In contrast, Rbm38-S195D protein was expressed as the slow-migrating Ser-195 phosphomimetic Rbm38 (Fig. 1, A and C, lanes 5 and 6). Interestingly, we found that both WT Rbm38 and S195A were capable of suppressing ΔNp63α expression in MCF7 and HaCaT cells (Fig. 1, A and C, lanes 1–4). In contrast, Rbm38-S195D did not suppress ΔNp63α expression in MCF7 cells (Fig. 1A, lanes 5 and 6) and even slightly increased ΔNp63α expression in HaCaT cells (Fig. 1C, lanes 5 and 6). Similar results were observed in ME180 and MIA-PaCa2 cells transiently transfected with a control vector or a vector expressing HA-tagged WT Rbm38, S195A, or S195D (Fig. S1). Consistent with this, we found that the level of p63α transcript was decreased by WT Rbm38 and S195A in MCF7 (Fig. 1B) and HaCaT cells (Fig. 1D). In contrast, upon expression of Rbm38-S195D, the level of p63α transcript was increased highly in HaCaT cells (Fig. 1D) and slightly in MCF7 cells (Fig. 1B).
Figure 1.
WT Rbm38 and S195A, but not S195D, inhibit the stability of p63 mRNA. A and C, levels of HA-tagged Rbm38, ΔNp63α, and actin proteins were measured in MCF7 cells (A) and HaCaT cells (C), both of which were uninduced or induced to express HA-tagged Rbm38, S195A, or S195D for 24 h. The relative ratio of p63 protein in the absence versus presence of Rbm38, S195A, or S195D is shown below each pair. B and D, levels of p63α and actin transcripts were measured in MCF7 cells (B) and HaCaT cells (D), both of which were uninduced or induced to express HA-tagged Rbm38, S195A, or S195D for 24 h. E, levels of Rbm38, p63, and actin proteins were measured in WT, Rbm38−/−, Rbm38S193A/S193A (A/A), and Rbm38 S193D/S193D (D/D) MEFs. F, levels of total p63, TAp63, and actin transcripts were measured in WT, Rbm38−/−, A/A, and D/D MEFs. G, half-life of p63 mRNA was determined in WT and D/D MEFs. The level of p63 transcript was measured by qRT-PCR in WT and D/D MEFs treated with DRB for various times.
To rule out potential nonphysiological artifacts of ectopic expression above, we generated Rbm38-KI mice in which Ser-193 (equivalent to Ser-195 in human Rbm38) is substituted with alanine (S193A) or aspartic acid (S193D). Next, these mice were used to generate a set of Rbm38S193A/S193A (A/A) and Rbm38S193D/S193D (D/D) MEFs along with WT and Rbm38−/− MEFs as a control. As shown in Fig. 1E, Rbm38 was detected in WT but not Rbm38−/− MEFs. We also showed that S193A and S193D proteins were expressed as two polypeptides with similar migration patterns as WT Rbm38 (Fig. 1E), suggesting that the slower migrating band of S193A and S193D proteins may be subjected to phosphorylation at other serine/threonine residues. Nevertheless, we found that loss of Rbm38 led to increased expression of p63 mRNA in Rbm38−/− MEFs as compared with that in WT MEFs (Fig. 1F, lanes 1 and 2), which is consistent with a previous report (12). Interestingly, we found that the level of p63 transcript was deceased in A/A MEFs but increased in D/D MEFs (Fig. 1F). These data indicate that the effect of ectopic Rbm38-S195A or Rbm38-S195D on p63 expression in MCF7 and HaCaT cells is recapitulated by endogenous knockin Rbm38-S193A and Rbm38-S193D in MEFs.
To determine whether the increased expression of p63 transcript by Rbm38-S193D is due to increased p63 mRNA stability, the half-life of p63 mRNA was determined in WT and D/D MEFs treated with 5,6-dichlorobenzimidazole-β-d-ribofuranoside (DRB; 100 μm) for various times. We found that the half-life of p63 mRNA was increased from 4.44 h in WT MEFs to 7.06 h in D/D MEFs (Fig. 1G). These data suggest that phosphomimetic S195D abrogates the ability of Rbm38 to decrease p63 mRNA stability.
Akt–GSK3 pathway modulates Ser-195 phosphorylation of Rbm38 and consequently p63 expression
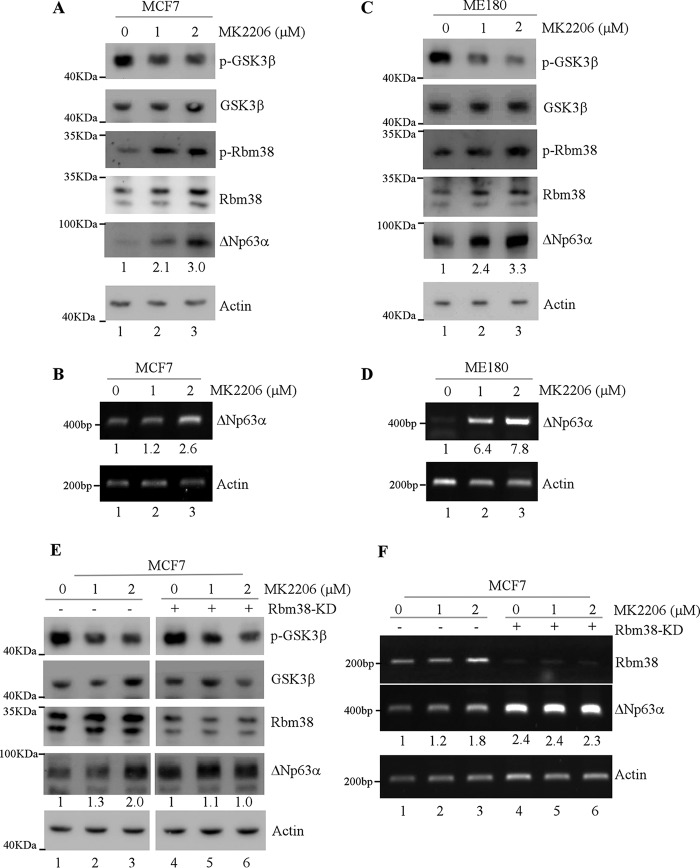
Glycogen synthase kinase-3β (GSK3β), a Ser/Thr kinase, is a master regulator for many cellular activities, including gene expression, cell cycle, and apoptosis (21). It is well known that Akt kinase phosphorylates GSK3β at Ser-9 and then inhibits GSK3β activity (22). Previously, we found that upon inhibition of the phosphatidylinositol 3-kinase (PI3K)–protein kinase B (Akt) pathway, GSK3β is activated and then phosphorylates Rbm38 at Ser-195, leading to increased p53 expression (20). Because Ser-195 phosphorylation alters the ability of Rbm38 to regulate p53 expression, we sought to determine whether p63 expression is regulated by the Akt–GSK3 pathway via Rbm38. To test this, MCF7 and ME180 cells were treated with MK2206, a potent inhibitor of Akt kinase (23). We showed that upon treatment with MK2206, the level of Ser-9–phosphorylated GSK3β, but not total GSK3β, was decreased in MCF7 and ME180 cells (Fig. 2, A and C, p-GSK3β and GSK3β panels), concomitantly with an increase in phosphorylated Rbm38 as detected by a specific antibody against Ser-195–phosphorylated Rbm38 (Fig. 2, A and C, p-Rbm38 panels). Importantly, we found that the levels of p63 protein and transcript were highly increased in MCF7 cells (Fig. 2, A and B) and ME180 cells (Fig. 2, C and D) by MK2206 in a dose-dependent manner.
Figure 2.
p63 expression is increased by Ser-195–phosphorylated Rbm38 via the Akt–GSK3 pathway. A and C, levels of p-GSK3β, GSK3β, p-Rbm38, Rbm38, p63α, and actin proteins were measured in MCF7 cells (A) and ME180 cells (C) treated with 0–2 μm MK2206 for 2 h. The relative level of ΔNp63α is shown below each lane. B and D, levels of ΔNp63 and actin proteins were measured in MCF7 cells (B) and ME180 cells (D) treated with 0–2 μm MK2206 for 2 h. The relative level of ΔNp63 transcript is shown below each lane. E, levels of p-GSK3β, GSK3β, Rbm38, p63α, and actin proteins were measured in MCF7 cells uninduced or induced to knock down Rbm38 for 48 h, followed by treatment with 0–2 μm MK2206 for 2 h. The relative level of p63α protein in mock-treated MCF7 cells (without or with Rbm38-KD) was arbitrarily set as 1.0, and the relative fold change is shown below each lane. F, levels of Rbm38, ΔNp63, and actin transcripts were measured in MCF7 cells uninduced or induced to knock down Rbm38 for 48 h, followed by treatment with 0–2 μm MK2206 for 2 h. The relative level of ΔNp63 transcript in mock-treated MCF7 cells was arbitrarily set as 1.0, and the relative fold change is shown below each lane.
To confirm that Rbm38 is necessary for enhanced p63 expression by the Akt inhibitor, Rbm38-knockdown MCF7 cells were treated with or without MK2206. We showed that the levels of p63 protein and transcript were increased in MCF7 cells upon treatment with MK2206 in a dose-dependent manner (Fig. 2, E and F, compare lanes 1 with 2 and 3). However, upon knockdown of Rbm38, MK2206 had little if any effect on the levels of p63 protein and transcript (Fig. 2, E and F, compare lanes 4 with 5 and 6). Additionally, we would like to note that the levels of p63 protein and transcript were increased in MCF7 cells upon knockdown of Rbm38 (Fig. 2, E and F, ΔNp63 panels), consistent with our previous study (13). These results suggest that p63 expression is increased by Ser-195 phosphorylation of Rbm38 via the Akt–GSK3 pathway.
U-rich element in p63 3′UTR is required for Rbm38 to regulate p63 expression
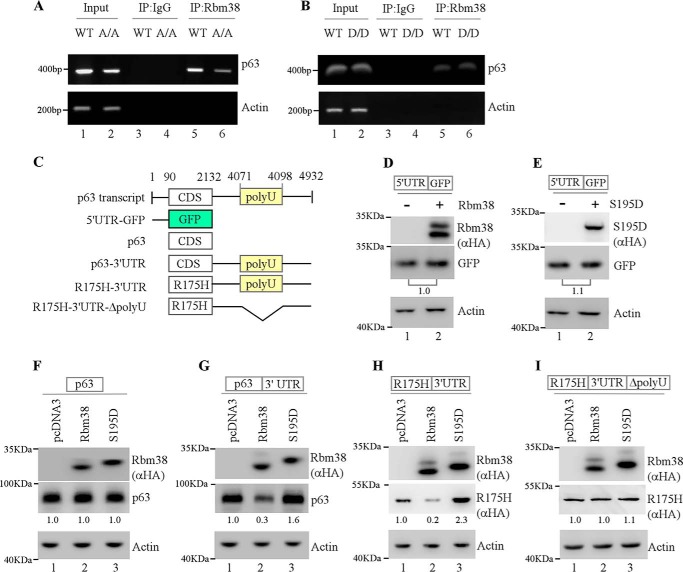
Previously, we reported that endogenous Rbm38 destabilizes p63 mRNA through binding to an AU/U-rich element in p63 3′UTR (13). To determine whether p63 3′UTR is recognized by S193D to regulate p63 expression, RNA ChIP assay was performed with anti-Rbm38 antibody using extracts from WT, A/A (Rbm38S193A/S193A) and D/D (Rbm38S193D/S193D) MEFs, followed by RT-PCR. IgG was used as an isotype control, and actin mRNA was measured as a negative control. We found that p63 mRNA was present in Rbm38/S193A/S193D, but not in control IgG, immunocomplexes (Fig. 3, A and B, p63 panels, compare lanes 3 and 4 with 5 and 6, respectively). These data suggest that like Rbm38, S193A and S193D are capable of binding to p63 mRNA. As a control, we found that actin mRNA was not detected in Rbm38, S193A, and S193D immunocomplexes (Fig. 3, A and B, Actin panel, compare lanes 3 and 4 with 5 and 6, respectively).
Figure 3.
U-rich element in p63 3′UTR is required for Rbm38 and S195D to regulate p63 expression. A and B, Rbm38, S193A, and S193D associate with p63 transcript. Cell lysates were collected from WT (A and B), Rbm38S193A/S193A (A), and Rbm38S193D/S193D (B) MEFs, followed by immunoprecipitation (IP) with anti-Rbm38 and rabbit IgG as a control. RT-PCR was performed to measure the levels of p63 and actin transcripts in Rbm38-, S193A-, or S193D–RNA complexes. C, schematic representation of p63 transcript and reporters. D and E, p63 5′UTR is not required for Rbm38 and S195D to regulate p63 expression. MCF7 cells were co-transfected with the p63 5′UTR-GFP reporter, along with a control vector, or a vector that expresses HA-tagged Rbm38 (D) or S195D (E) for 24 h. Cell lysates were collected and subjected to Western blot analysis to determine the levels of HA-tagged Rbm38, S195D, GFP, and actin proteins. The level of GFP protein was normalized to that of actin, and the relative ratio of GFP protein is shown below each pair. F and G, p63 3′UTR is required for Rbm38 to regulate p63 expression. MCF7 cells were co-transfected with a reporter containing p63 coding region alone (F) or together with p63 3′UTR (G), along with a control vector or a vector that expresses HA-tagged Rbm38 or S195D. Cell lysates were collected and subjected to Western blot analysis to determine the levels of HA-tagged Rbm38, HA-S195D, p63, and actin proteins. The level of p63 protein was normalized to that of actin, and the relative fold change is shown below each lane. H and I, U-rich element in p63 3′UTR is required for Rbm38 to regulate p63 expression. MCF7 cells were co-transfected with the p53-R175H reporter containing an intact p63 3′UTR (H) or poly(U)-deleted p63 3′UTR (I), along with a control vector or a vector that expresses HA-tagged Rbm38 or S195D for 24 h. Cell lysates were collected and subjected to Western blot analysis to determine the levels of HA-Rbm38, HA-S195D, HA-p53-R175H, and actin proteins. The level of HA-p53-R175H was normalized to that of actin, and the relative fold change is shown below each lane.
Next, to determine the Rbm38-binding site in p63α 3′UTR, multiple reporters were generated (Fig. 3C). First, a reporter carrying the GFP-coding region plus p63 5′UTR (Fig. 3, D and E) was transfected into MCF7 cells together with an empty vector or a vector expressing HA-tagged Rbm38 or Rbm38-S195D. We found that both Rbm38 and S195D had no effect on the level of GFP from a reporter that carries the GFP-coding region plus p63 5′UTR (Fig. 3, D and E, GFP panel, compare lanes 1 and 2). Next, a reporter that carries the p63-coding region alone or together with p63 3′UTR was used. We found that neither Rbm38 nor S195D had an effect on the p63 expression from a p63 reporter that carries only the p63-coding region (Fig. 3F). Interestingly, we found that the level of p63 protein was repressed by Rbm38, but increased by S195D, from the reporter that carries the p63-coding region plus p63 3′UTR (Fig. 3G, compare lane 1 with 2 and 3, respectively). To verify this, we generated a reporter, which carries HA-tagged mutant p53-R175H along with p63 3′UTR (Fig. 3C). Consistently, we found that the level of p53-R175H was decreased by Rbm38 but increased by S195D (Fig. 3H). Rbm38 is known to bind to p63 mRNA via a poly(U)-rich element (13). Thus, we generated another reporter, which carries HA-tagged mutant p53-R175H along with poly(U)-deleted p63 3′UTR (lacking nt 4071–4098) (Fig. 3C). We found that upon deletion of the poly(U)-rich element both Rbm38 and S195D were unable to regulate p53-R175H expression (Fig. 3I). These data suggest that the poly(U)-rich element in p63 3′UTR is necessary for Rbm38 and S195D to regulate p63 mRNA stability.
Rbm38 but not Rbm38-S195D recruits miR203 to target p63 mRNA for degradation
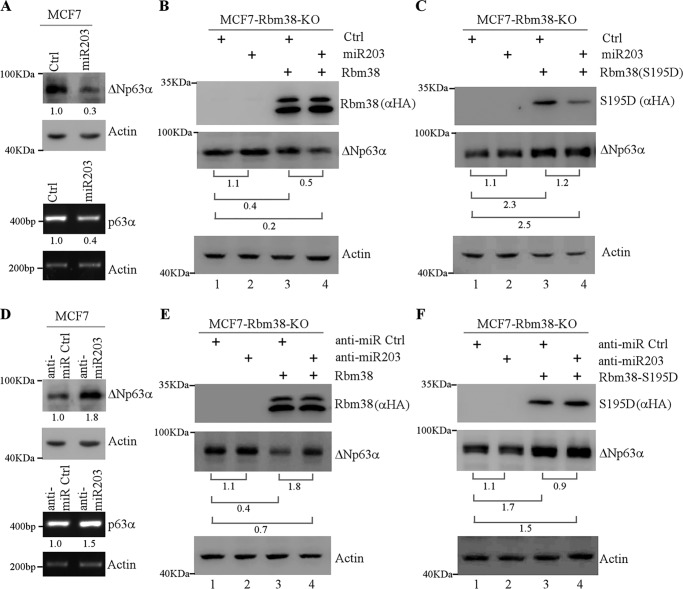
miRNAs are known to regulate RNA stability by binding to the 3′UTR of a target mRNA (24–27). miR203 has been identified as a skin-specific microRNA, and previous studies showed that miR203 regulates p63 expression by directly binding to p63 3′UTR (28, 29). Interestingly, the binding site for miR203 in p63 3′UTR is adjacent to the one for Rbm38. Because Rbm38 is known to modulate the access of miRNAs to its targets (16), we wanted to determine whether miR203 is involved in the differential regulation of p63 mRNA stability by Rbm38 and S195D. To test this, multiple cell lines were used and transfected with a control miRNA or miR203 mimic. miR203 mimic is a small, chemically modified dsRNA that mimics endogenous miR203. We showed that the levels of p63 protein and transcript were decreased by miR203 in MCF7 cells (Fig. 4A, top and bottom panels) and ME180 and Mia-PaCa2 cells (Fig. S2, A and B), consistent with the previous study (28). Next, to determine whether Rbm38 is involved in miR203-mediated p63 expression, we generated the Rbm38-KO MCF7 cell line by using CRISPR/Cas9 technology. We showed that in the absence of Rbm38, miR203 had very little effect on p63 expression (Fig. 4B, p63 panel, compare lanes 1 and 2). Upon ectopic expression of HA-tagged Rbm38, p63 expression was markedly inhibited by Rbm38 (Fig. 4B, p63 panel, compare lanes 1 and 3), which was further inhibited by co-expression of miR203 (Fig. 4B, p63 panel, compared lane 3 with 4; also compare lanes 1 and 4). These data suggest that the ability of miR203 to suppress p63 expression is Rbm38-dependent. In contrast, p63 expression was increased by S195D in Rbm38-KO cells (Fig. 4C, compare lanes 1 and 3). Moreover, miR203 had no effect on the ability of S195D to increase p63 expression (Fig. 4C, compare lanes 3 and 4).
Figure 4.
Rbm38 but not Rbm38-S195D recruits miR203 to target p63 mRNA for degradation. A, MCF7 cells were transfected with a control miRNA or miR203 mimic for 48 h. The levels of p63 and actin proteins (upper panel) and transcripts (lower panel) were measured in MCF7 cells. The relative level of p63 protein and transcript is shown below each lane. B and C, Rbm38-KO MCF7 cells were transiently transfected with a control (Ctrl) vector or a vector that expresses HA-tagged Rbm38 (B) or HA-S195D (C), along with a control miRNA or miR203 mimic for 48 h. Cell lysates were collected and subjected to Western blot analysis with anti-HA (HA-Rbm38 or HA-S195D), anti-p63, and anti-actin. The relative level of p63 was determined, and the relative ratio is shown below each pair. D, MCF7 cells were transfected with a control anti-miRNA or anti-miR203 for 48 h. The levels of p63 protein and actin proteins and transcripts were measured in MCF7 cells. The relative level of p63 is shown below each lane. E and F, Rbm38-KO MCF7 cells were transiently transfected with a control vector or a vector that expresses HA-tagged Rbm38 (E) or HA-S195D (F), along with a control anti-miRNA or anti-miR203 for 48 h. Cell lysates were collected and subjected to Western blot analysis with anti-HA (HA-Rbm38 or HA-S195D), anti-p63, and anti-actin. The relative level of p63 was determined, and the relative ratio was shown below each pair.
To confirm the above observation, miR203 inhibitor (anti-miR-203), which specifically inhibits endogenous miR203, was used to examine its effect on p63 expression. We found that the levels of p63 protein and transcript were increased by anti-miR203 in MCF7 cells (Fig. 4D, top and lower panels) and ME180 and Mia-PaCa2 cells (Fig. S2, C and D), consistent with the previous report (28). Next, Rbm38-KO MCF7 cells were transiently transfected with a control or anti-miR203 along with or without Rbm38 expression. We found that in the absence of Rbm38, anti-miR203 had very little effect on p63 expression in Rbm38-KO MCF7 cells (Fig. 4E, compare lanes 1 and 2). In the presence of Rbm38, anti-miR203 was able to counter the inhibition of p63 expression by Rbm38 (Fig. 4E, compare lanes 3 and 4; also compare lanes 1 and 4). By contrast, we found that anti-miR203 was not able to further elevate p63 expression increased by S195D (Fig. 4F, compare lanes 3 and 4), which is consistent with the above observation that miR203 did not abrogate the ability of S195D to increase p63 expression (Fig. 4C, compare lanes 3 and 4).
Rbm38 recruits Argonaute2 (Ago2) for miR203-mediated degradation of p63 transcript
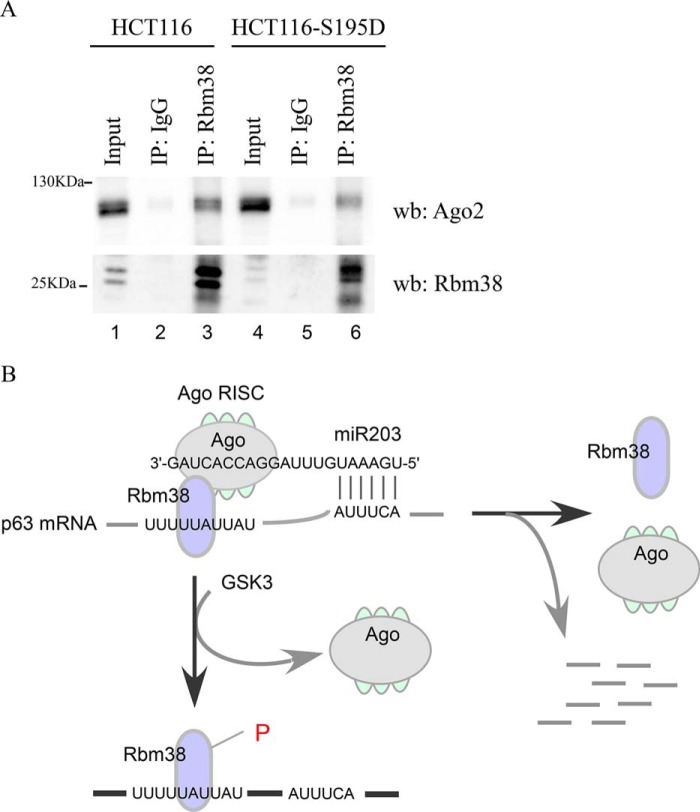
Previously, Rbm38 was found to modulate the ability of several miRNAs to target their RNA substrates Ago2 (16), which is known to be required for miRNA-mediated mRNA decay (30). Additionally, we found that miR203 was capable of inhibiting p63 expression in the presence of Rbm38 but not S195D (Fig. 4, B and C and E and F). This let us speculate that S195D prevents miR203 from binding to the p63 mRNA, leading to enhanced p63 expression. To test this, we generated HCT116-S195D stable cell lines in that endogenous Ser-195 in Rbm38 was substituted with aspartic acid. Next, HCT116-S195D cells along with isogenic control cells were used to examine the association of endogenous Ago2 with endogenous Rbm38 or Rbm38-S195D by immunoprecipitation followed by Western blot analysis. We found that Ago2 was detectable in Rbm38 immunocomplexes in HCT116 cells (Fig. 5A, lane 3). However, the association between Ago2 and Rbm38-S195D was weaker in HCT116-S195D cells (Fig. 5A, lane 6), suggesting that phosphorylation of Ser-195 reduces the association between Rbm38 and Ago2. Together, these data suggest that the association between Rbm38 and Ago2 is required for miR203-mediated degradation of p63 transcript.
Figure 5.
Phosphorylation of Ser-195 weakens the association between Rbm38 and Ago2. A, cell lysates from isogenic control and S195D-KI HCT116 cells were immunoprecipitated (IP) with control IgG, anti-Rbm38, or anti-Ago2, followed by Western blot (wb) analysis with antibodies against Rbm38 and Ago2. B, model for how Ser-195 phosphorylation modulates the ability of Rbm38 to regulate p63 expression.
Discussion
Although Rbm38 regulates both p53 and p63 expression, the mechanisms by which p53 and p63 are regulated differently (13, 15, 20). Previously, we found that Rbm38 binds to p63 3′UTR and inhibits p63 expression via decreased mRNA stability (13). In contrast, Rbm38 interacts with eIF4E and represses eIF4E from binding to p53 5′UTR, leading to inhibition of p53 mRNA translation (15). Interestingly, Ser-195 phosphorylation of Rbm38 abrogates the interaction between Rbm38 and eIF4E, resulting in an increased p53 translation (20). In this study, we found that phosphomimetic S195D abrogates the ability of Rbm38 to decrease p63 mRNA stability (Fig. 1). Additionally, activation of GSK3 leads to increased Rbm38 phosphorylation along with increased p63 expression in an Rbm38-dependent manner (Fig. 2). Mechanistically, we found that phosphomimetic S195D attenuates the association of Rbm38 with Ago2, which prevents miR203 from targeting p63 mRNA for degradation. Together, we uncovered a novel mechanism by which Ser-195 phosphorylation modulates its ability of Rbm38 to regulate p63 mRNA stability via miR203 as summarized in Fig. 5B.
Rbm38 is known to recognize AU/U-rich elements, but the precise region to which Rbm38 binds has not been located in the p63 transcript. Here, using a reporter assay and mutational analysis, a U-rich element (nt 4071–4098) in p63 3′UTR is found to be recognized by Rbm38 and necessary for Rbm38 to regulate p63 mRNA stability (Fig. 3). It is of note that RNA-binding proteins often modulate miRNA activity by enhancing or blocking the binding of miRNAs to their targets (31). Indeed, Rbm38 is found to selectively inhibit the accessibility of miRNAs to the 3′UTRs of p21, LATS2, and Sirt1 transcripts (16). Although the p63 transcript is recognized and regulated by miR203 (28, 32, 33), it remains unclear whether Rbm38 and miR203 are coordinated to regulate p63 mRNA stability. Here, we showed that Rbm38 is required for miR203 to target p63 mRNA for degradation (Figs. 3 and 4). Interestingly, phosphomimetic S195D attenuates Rbm38 from interacting with Ago2 and thus reduces miR203 to target p63 mRNA for degradation, although Rbm38-S195D is still capable of binding to p63 3′UTR (Figs. 3–5). These observations prompted us to speculate that upon activation of GSK3 kinases and/or inhibition of Akt kinases, Rbm38 is phosphorylated at Ser-195, which would attenuate the ability of Rbm38 to interact with Ago2 as well as the ability of Rbm38 to recruit miR203 to target p63 mRNA for degradation (Fig. 5B).
Our previous study showed that GSK3β phosphorylates Rbm38, which is consistent with a previous report that GSK3β is activated by endoplasmic reticulum stress to phosphorylate p53 (20, 34) and by DNA damage to phosphorylate TIP60 (35). In this study, we found that blocking the PI3K–Akt pathway leads to activation of the GSK3β kinase, which in turn increases p63 expression in an Rbm38-dependent manner, consistent with the report that inhibition of GSK3β kinase activity decreases ΔNp63α expression under a nonstress condition (36). Thus, further understanding of how GSK3β regulates Rbm38 phosphorylation and p63 activity under both basal and stress conditions is warranted, especially considering that GSK3 inhibitors are currently being explored as a therapeutic agent for cancer and cardiovascular and neurodegenerative diseases.
As an inhibitor of p63 mRNA stability, Rbm38 can be targeted to modulate p63 expression. Indeed, we found recently that compound Rbm38−/−;TAp63+/− mice, wherein the Rbm38 deficiency restores p63 expression to near normal levels, have a longer life span along with reduced tumor incidence as compared with TAp63+/− or Rbm38−/− mice (12). Given that Ser-195 phosphorylation of Rbm38 enhances p63 expression, future experiments are warranted to determine whether Rbm38 phosphorylation and/or phosphomimetic S195D modulates the ability of Rbm38 to regulate p63-dependent tumor suppression and premature aging.
Materials and methods
Reagents
Proteinase inhibitor mixture, RNase A, 5,6-dichlorobenzimidazole-β-d-ribofuranoside, and protein A/G beads were purchased from Sigma. MK2206 was purchased from SelleckChem (Houston, TX). RevertAid First Strand cDNA Synthesis kit was purchased from ThermoFisher ScientificTM (Carlsbad, CA).
Plasmids
pcDNA4 and pcDNA3 vectors expressing HA-tagged Rbm38, HA-tagged Rbm38-S195A, or HA-tagged Rbm38-S195D were generated as described previously (20). The pcDNA3-TAp63-CDS and pcDNA3-TAp63–3′UTR expression vectors were generated as described previously (37). pcDNA3 vector expressing GFP was generated as described previously (20). pcDNA3 vector expressing HA-tagged p53-R175H was generated as described previously (14). The pcDNA3 p53-R175H-3′UTR reporter was generated by cloning p63 3′UTR into pcDNA3-HA-p53-R175H downstream of p53-R175H-CDS. p63 3′UTR was amplified with forward primer, 5′-GGG CTC GAG GCC TCA CCA TGT GAG CTC TTC C-3′, and reverse primer, 5′-TCT AGA GCA TGT CCT GGC AAA CAA AAA G-3′. To generate pcDNA3 p53-R175H-3′UTR-Δpoly (U), which lacks a U-rich region (nt 4071–4098), a two-step PCR strategy was used. The first-step PCR was performed to separately amplify two DNA fragments by using pcDNA3-p63–3′UTR as a template. Fragment 1 was amplified with forward primer, 5′-GGG CTC GAG GCC TCA CCA TGT GAG CTC TTC C-3′, and reverse primer, 5′-CTC ATT CTC TTT AAC ATA CCT TTC CCT TCC CTC-3′. Fragment 2 was amplified with forward primer, 5′-GAA GGG AAA GGG TAT GTT AAA GAG AAT-3′, and reverse primer, 5′-TCT AGA GCA TGT CCT GGC AAA CAA AAA G-3′. The second-step PCR was performed using a mixture of fragments 1 and 2 as a template with forward primer, 5′-GGG CTC GAG GCC TCA CCA TGT GAG CTC TTC C-3′, and reverse primer, 5′-TCT AGA GCA TGT CCT GGC AAA CAA AAA G-3′. This PCR product was then inserted into pcDNA3-HA-p53-R175H vector via BamHI and XhoI sites to generate pcDNA3-HA-p53-R175H-3′UTR-Δpoly (U). Rbm38 guide RNAs (gRNAs) were designed using CRISPR design tool. To generate a vector expressing a single-guide RNA (sgRNA) targeting Rbm38, two 25-nt oligonucleotides were annealed and then cloned into pSpCas9 (BB) sgRNA expression vector (41). The sgRNA sequence for Rbm38-KO is 5′-ACA CTA CCG ACG CCT CGC TC-3′. The sgRNA sequence for Rbm38-S195D-KI is 5′-CGA GGC GGC GTA TGG GTA G-3′. To generate the S195D-KI HR template, an 800-bp DNA fragment was amplified using HCT116 genomic DNA with a forward primer, 5′-GGC GTA TGG GTA CTG GTC AT-3′, and a reverse primer, 5′-GAA TGC CCG AGC GGT CTT CG-3′. This DNA fragment was then cloned into pGEMT vector and used as a template for two-step PCR. The first step was to amplify two DNA fragments #1 and #2. The DNA fragment #1 was amplified using forward primer #1, 5′-GGC GTA TGG GTA CTG GTC AT-3′, and reverse primer #1, 5′CTG TAG CCC ACG AAC GAC GCA GCC GTG GCA GGA TCC GCG GCG TAT GGG TAC TGA TCA TAT GTG GCC GG-5′. The DNA fragment #2 was amplified using forward primer #2, 5′-CCG GCC ACA TAT GAT CAG TAC CCA TAC GCC GCG GAT CCT GCC ACG GCT GCG TCG TTC GTG GGC TAC AG-3′, and reverse primer #2, 5′-GAA TGC CCG AGC GGT CTT CG-3′. The second-step PCR was amplified using DNA fragments #1 and #2 as template along with forward primer #1 and reverse primer #2. The PCR product was then cloned to the pGEMT vector, and the S195D-KI template was confirmed by sequencing. Next, the S195D-KI vector was used as a template to perform asymmetric PCR only using forward primer #1, and the resulting PCR products were purified and then used as HR template for knockin cell generation.
Cell culture
Rbm38+/+, Rbm38−/−, Rbm38S193A/S193A, and Rbm38S193D/S193D MEFs were generated as described previously (11). Rbm38-S193A and Rbm38-S193D mice were generated by HR strategy (42) HaCaT cells expressing HA-tagged Rbm38, Rbm38-S195A, Rbm38-S195D were generated as described previously (20). Rbm38-KO MCF7 cell lines were generated by using CRISPR/Cas9 technology, as described previously (38). HCT116-S195D cells were generated by using CRISPR-Cas9 technology. Briefly, HCT116 cells were transfected with Rbm38-KI gRNA vector and Rbm38-KI HR template. Cells were selected with puromycin for 3 weeks. Individual clones were picked and then confirmed by sequence analysis. MCF7, HaCaT, ME180, Mia-PaCa2, HCT116, and their derivatives were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT).
RNA isolation, RT-PCR, and quantitative RT-PCR
Total RNA was isolated with TRIzol reagent (Invitrogen). cDNA was synthesized using RevertAid First Strand cDNA Synthesis kit according to the manufacturer's protocol (ThermoFisher ScientificTM). The primers used to amplify actin (human) were forward primer, 5′-CTG AAG TAC CCC ATC GAG CAC GGC A-3′, and reverse primer, 5′-GGA TAG CAC AGC CTG GAT AGC AAC G-3′. The primers used to amplify human p63 were forward primer, 5′-CAG CAT GGA CCA GCA GAT TC-3′, and reverse primer, 5′-GGT CAT CAC CTT GAT CTG GA-3′. The primers used to amplify human TAp63 were forward primer, 5′-TTA TTA CCG ATC CAC CAT GTC-3′, and reverse primer, 5′-TGC GGA TAC AGT CCA TGC TA-3′. The primers used to amplify human ΔNp63 were forward primer, 5′-AAG GAA ATG AAT TTT GA-3′, and reverse primer, 5′-TGC GGA TAC AGT CCA TGC TA-3′. The primers used to amplify human p63α were forward primer, 5′-CAT GAA CAA GCT GCC TTC TG-3′, and reverse primer, 5′-AGG AGA ATT GGT GGA GCT G-3′. The primers used to amplify human Rbm38 were forward primer, 5′-CT ACC GAC GCC TCG CTC AG-3′, and reverse primer, 5′-CCC AGA TAT GCC AGG TTC AC-3′. The primers used to amplify murine p63 were forward primer, 5′-GAA GAG ACC GGA AGG CAG AT-3′, and reverse primer, 5′-CAT CAT CTG GGG ATC TCC GT-3′. The primers used to amplify murine TAp63 were forward primer, 5′-TAC AGA TCT GCC ATG TCG CA-3′, and reverse primer, 5′-GCA TGC GGA TAC AAT CCA TG-3′.
For qRT-PCR analysis, 15-μl reactions were set up using 2× qPCR SYBR Green Mix (ABgene) along with 5 μmol/liter primers. The reactions were run on a StepOne plus (Invitrogen) using a two-step cycling program: 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 68 °C for 30 s. A melt curve (57–95 °C) was generated at the end of each run to verify the specificity. The primers for murine p63 were forward primer, 5′-GAA GAG ACC GGA AGG CAG AT-3′, and reverse primer, 5′-CAT CAT CTG GGG ATC TCC GT-3′. The primers for mouse actin were forward primer, 5′-CCC ATC TAC GAG GGC TAT-3′, and reverse primer, 5′-AGA AGG AAG GCT GGA AAA-3′.
mRNA half-life assay
To measure the stability of p63 mRNA, WT or D/D MEFs were treated with 5,6-dichlorobenzimidazole-β-d-ribofuranoside (DRB; 100 μm) for various times. The relative level of p63 mRNA was determined by qRT-PCR and then normalized to the level of actin mRNA from three separate experiments. The half-life of p63 mRNA was plotted versus time.
Western blot analysis and immunoprecipitation–Western blot analysis
Western blotting was performed as described previously (39). Briefly, cell lysates was resolved in 8–12% SDS-polyacrylamide gel and then transferred to nitrocellulose membrane. The blots were then incubated with a primary and then a secondary antibody, followed by detection with enhanced chemiluminescence. To perform the IP-WB analysis, cells were lysed in 1.0% Triton lysis buffer (50 mm Tris (pH 7.4), 150 mm NaCl, 1.0% Triton X-100) supplemented with the proteinase inhibitor mixture (100 μg/ml), followed by incubation with 1 μg of antibody or control IgG. The immunocomplexes were brought down by protein A/G beads and subjected to Western blot analysis. Antibodies against p63 (4A4) and GFP were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-GSK3β, anti-p-GSK3β (specific to serine 9), and anti-Ago2 were purchased from Cell Signaling Technology (Beverly, MA). Anti-Rbm38 antibody was made as described previously (20). Anti-phospho-Ser-195 Rbm38 antibody (anti-p-Rbm38) was generated by OpenBiosystems by using a phosphopeptide spanning serine 195 (188DQYPYAAS(p)PATA199). Anti-HA was purchased from Covance (San Diego). Anti-actin and horseradish peroxidase-conjugated secondary antibodies against rabbit and mouse IgG were purchased from Bio-Rad. The immunoreactive bands were visualized by enhanced chemiluminescence (ThermoFisher Scientific) and quantified by densitometry with the BioSpectrum 810 Imaging System (UVP LLC, Upland, CA).
RNA-IP assay
RNA immunoprecipitation was carried out as described previously (40). Briefly, cells extracts were prepared with immunoprecipitation buffer (10 mm HEPES, pH 7.0, 100 mm KCl, 5 mm MgCl2, 0.5% Nonidet P-40, and 1 mm DTT) and then incubated with 2 μg of anti-Rbm38 or an isotype control IgG overnight at 4 °C. The RNA–protein immunocomplexes were brought down by protein A beads. RT-PCR analysis was carried out to determine the levels of p63 and actin transcripts.
Micro-RNA transfection
Micro-RNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. The miRNAs used were mirVanaTM miRNA negative control and miR203 mimic (ID: MC10152, Ambion–ThermoFisher Scientific) with a sequence of 5′-GUG AAA UGU UUA GGA CCA CUA G-3′ (Double-stranded RNA). miRNA inhibitors (anti-miRNA) used in this study were mirVanaTM miRNA control and miR203-specific inhibitor (ID: MH10152, Ambion–ThermoFisher Scientific) with a sequence of 5′-GUG AAA UGU UUA GGA CCA CUA G-3′ (single-stranded RNA).
Author contributions
Y. Z. and X. C. data curation; Y. Z. and X. C. formal analysis; Y. Z. validation; Y. Z., X. F., and W. S. methodology; Y. Z. writing-original draft; Y. Z., J. Z., and X. C. writing-review and editing; X. C. supervision; X. C. funding acquisition; X. C. project administration.
Supplementary Material
This work is supported in part by National Institutes of Health Grant CA195828. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
- miRNA
- microRNA
- MEF
- mouse embryo fibroblast
- DRB
- 5,6-dichlorobenzimidazole-β-d-ribofuranoside
- GSK3β
- glycogen synthase kinase-3β
- PI3K
- phosphatidylinositol 3-kinase
- Akt
- protein kinase B
- sgRNA
- single-guide RNA
- gRNA
- guide RNA
- IP
- immunoprecipitation
- nt
- nucleotide.
References
- 1. Levrero M., De Laurenzi V., Costanzo A., Gong J., Wang J. Y., and Melino G. (2000) The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci. 113, 1661–1670 [DOI] [PubMed] [Google Scholar]
- 2. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., and McKeon F. (1998) p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316 10.1016/S1097-2765(00)80275-0 [DOI] [PubMed] [Google Scholar]
- 3. Osada M., Ohba M., Kawahara C., Ishioka C., Kanamaru R., Katoh I., Ikawa Y., Nimura Y., Nakagawara A., Obinata M., and Ikawa S. (1998) Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 4, 839–843 10.1038/nm0798-839 [DOI] [PubMed] [Google Scholar]
- 4. Koster M. I., Kim S., Mills A. A., DeMayo F. J., and Roop D. R. (2004) p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 18, 126–131 10.1101/gad.1165104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., and Bradley A. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- 6. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., and McKeon F. (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718 10.1038/19539 [DOI] [PubMed] [Google Scholar]
- 7. Feldstein O., Ben-Hamo R., Bashari D., Efroni S., and Ginsberg D. (2012) RBM38 is a direct transcriptional target of E2F1 that limits E2F1-induced proliferation. Mol. Cancer Res. 10, 1169–1177 10.1158/1541-7786.MCR-12-0331 [DOI] [PubMed] [Google Scholar]
- 8. Shu L., Yan W., and Chen X. (2006) RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 20, 2961–2972 10.1101/gad.1463306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu E., Zhang J., and Chen X. (2013) MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability. Oncogene 32, 2169–2178 10.1038/onc.2012.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan W., Zhang J., Zhang Y., Jung Y. S., and Chen X. (2012) p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability. Mol. Cell. Biol. 32, 2336–2348 10.1128/MCB.00215-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J., Xu E., Ren C., Yang H. J., Zhang Y., Sun W., Kong X., Zhang W., Chen M., Huang E., and Chen X. (2018) Genetic ablation of Rbm38 promotes lymphomagenesis in the context of mutant p53 by downregulating PTEN. Cancer Res. 78, 1511–1521 10.1158/0008-5472.CAN-17-2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang Y., Xu E., Zhang J., Chen M., Flores E., and Chen X. (2018) The Rbm38-p63 feedback loop is critical for tumor suppression and longevity. Oncogene 37, 2863–2872 10.1038/s41388-018-0176-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J., Jun Cho S., and Chen X. (2010) RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc. Natl. Acad. Sci. U.S.A. 107, 9614–9619 10.1073/pnas.0912594107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan W., Zhang Y., and Chen X. (2017) TAp63γ and ΔNp63γ are regulated by RBM38 via mRNA stability and have an opposing function in growth suppression. Oncotarget 8, 78327–78339 10.18632/oncotarget.18463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J., Cho S. J., Shu L., Yan W., Guerrero T., Kent M., Skorupski K., Chen H., and Chen X. (2011) Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 25, 1528–1543 10.1101/gad.2069311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Léveillé N., Elkon R., Davalos V., Manoharan V., Hollingworth D., Oude Vrielink J., le Sage C., Melo C. A., Horlings H. M., Wesseling J., Ule J., Esteller M., Ramos A., and Agami R. (2011) Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat. Commun. 2, 513 10.1038/ncomms1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobs K. M., Bhave S. R., Ferraro D. J., Jaboin J. J., Hallahan D. E., and Thotala D. (2012) GSK-3β: a bifunctional role in cell death pathways. Int. J. Cell Biol. 2012, 930710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Daugherty R. L., and Gottardi C. J. (2007) Phospho-regulation of β-catenin adhesion and signaling functions. Physiology 22, 303–309 10.1152/physiol.00020.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., and He X. (2002) Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847 10.1016/S0092-8674(02)00685-2 [DOI] [PubMed] [Google Scholar]
- 20. Zhang M., Zhang J., Chen X., Cho S. J., and Chen X. (2013) Glycogen synthase kinase 3 promotes p53 mRNA translation via phosphorylation of RNPC1. Genes Dev. 27, 2246–2258 10.1101/gad.221739.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kockeritz L., Doble B., Patel S., and Woodgett J. R. (2006) Glycogen synthase kinase-3–an overview of an over-achieving protein kinase. Curr. Drug Targets 7, 1377–1388 10.2174/1389450110607011377 [DOI] [PubMed] [Google Scholar]
- 22. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., and Hemmings B. A. (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 10.1038/378785a0 [DOI] [PubMed] [Google Scholar]
- 23. Hirai H., Sootome H., Nakatsuru Y., Miyama K., Taguchi S., Tsujioka K., Ueno Y., Hatch H., Majumder P. K., Pan B. S., and Kotani H. (2010) MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol. Cancer Ther. 9, 1956–1967 10.1158/1535-7163.MCT-09-1012 [DOI] [PubMed] [Google Scholar]
- 24. Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., and Pasquinelli A. E. (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 10.1016/j.cell.2005.07.031 [DOI] [PubMed] [Google Scholar]
- 25. Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., and Izaurralde E. (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898 10.1101/gad.1424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., Inoue K., Enright A. J., and Schier A. F. (2006) Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- 27. Wu L., Fan J., and Belasco J. G. (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U.S.A. 103, 4034–4039 10.1073/pnas.0510928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yi R., Poy M. N., Stoffel M., and Fuchs E. (2008) A skin microRNA promotes differentiation by repressing 'stemness'. Nature 452, 225–229 10.1038/nature06642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kouwenhoven E. N., Oti M., Niehues H., van Heeringen S. J., Schalkwijk J., Stunnenberg H. G., van Bokhoven H., and Zhou H. (2015) Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 16, 863–878 10.15252/embr.201439941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okamura K., Ishizuka A., Siomi H., and Siomi M. C. (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18, 1655–1666 10.1101/gad.1210204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., and Filipowicz W. (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 10.1016/j.cell.2006.04.031 [DOI] [PubMed] [Google Scholar]
- 32. Truong A. B., and Khavari P. A. (2007) Control of keratinocyte proliferation and differentiation by p63. Cell Cycle 6, 295–299 10.4161/cc.6.3.3753 [DOI] [PubMed] [Google Scholar]
- 33. Viticchiè G., Lena A. M., Latina A., Formosa A., Gregersen L. H., Lund A. H., Bernardini S., Mauriello A., Miano R., Spagnoli L. G., Knight R. A., Candi E., and Melino G. (2011) MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 10, 1121–1131 10.4161/cc.10.7.15180 [DOI] [PubMed] [Google Scholar]
- 34. Qu L., Huang S., Baltzis D., Rivas-Estilla A. M., Pluquet O., Hatzoglou M., Koumenis C., Taya Y., Yoshimura A., and Koromilas A. E. (2004) Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3β. Genes Dev. 18, 261–277 10.1101/gad.1165804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Charvet C., Wissler M., Brauns-Schubert P., Wang S. J., Tang Y., Sigloch F. C., Mellert H., Brandenburg M., Lindner S. E., Breit B., Green D. R., McMahon S. B., Borner C., Gu W., and Maurer U. (2011) Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol. Cell 42, 584–596 10.1016/j.molcel.2011.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu J., Liang S., Bergholz J., He H., Walsh E. M., Zhang Y., and Xiao Z. X. (2014) ΔNp63α activates CD82 metastasis suppressor to inhibit cancer cell invasion. Cell Death Dis. 5, e1280 10.1038/cddis.2014.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu E., Zhang J., Zhang M., Jiang Y., Cho S. J., and Chen X. (2014) RNA-binding protein RBM24 regulates p63 expression via mRNA stability. Mol. Cancer Res. 12, 359–369 10.1158/1541-7786.MCR-13-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y., Qian Y., Zhang J., Yan W., Jung Y. S., Chen M., Huang E., Lloyd K., Duan Y., Wang J., Liu G., and Chen X. (2017) Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev. 31, 1243–1256 10.1101/gad.299388.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Yan W., and Chen X. (2014) p63 regulates tubular formation via epithelial-to-mesenchymal transition. Oncogene 33, 1548–1557 10.1038/onc.2013.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peritz T., Zeng F., Kannanayakal T. J., Kilk K., Eiríksdóttir E., Langel U., and Eberwine J. (2006) Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 1, 577–580 10.1038/nprot.2006.82 [DOI] [PubMed] [Google Scholar]
- 41. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rouet P., Smih F., and Jasin M. (1994) Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 91, 6064–6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.